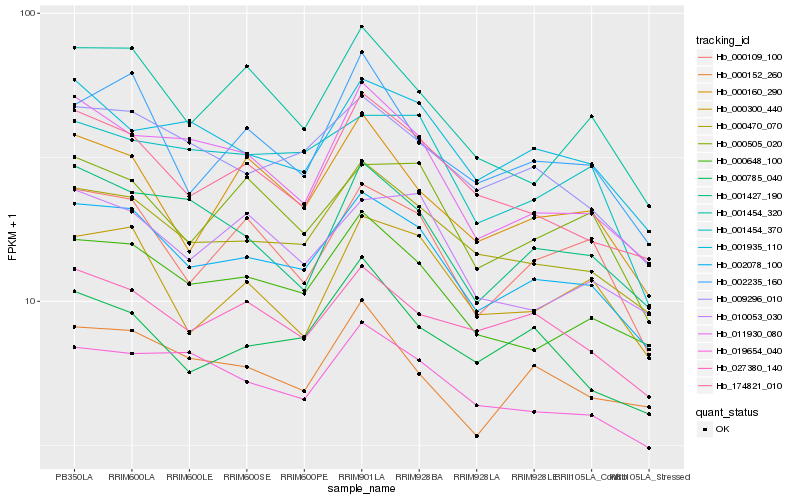
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_100 |
0.0 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 2 |
Hb_000470_070 |
0.0425734844 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 3 |
Hb_009296_010 |
0.0529674484 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 4 |
Hb_000648_100 |
0.0543203235 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 5 |
Hb_174821_010 |
0.0614129022 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 6 |
Hb_000109_100 |
0.0634064317 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 7 |
Hb_019654_040 |
0.0647103396 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001935_110 |
0.0656085458 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 9 |
Hb_027380_140 |
0.0671811716 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011930_080 |
0.0676698755 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 11 |
Hb_000505_020 |
0.0676862272 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 12 |
Hb_001427_190 |
0.0703366708 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 13 |
Hb_001454_320 |
0.0711856628 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010053_030 |
0.0715180777 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 15 |
Hb_000160_290 |
0.0729379464 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001454_370 |
0.0732894522 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 17 |
Hb_000152_260 |
0.0734347801 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 18 |
Hb_002235_160 |
0.074099987 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 19 |
Hb_000300_440 |
0.0748255655 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000785_040 |
0.0749548125 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |