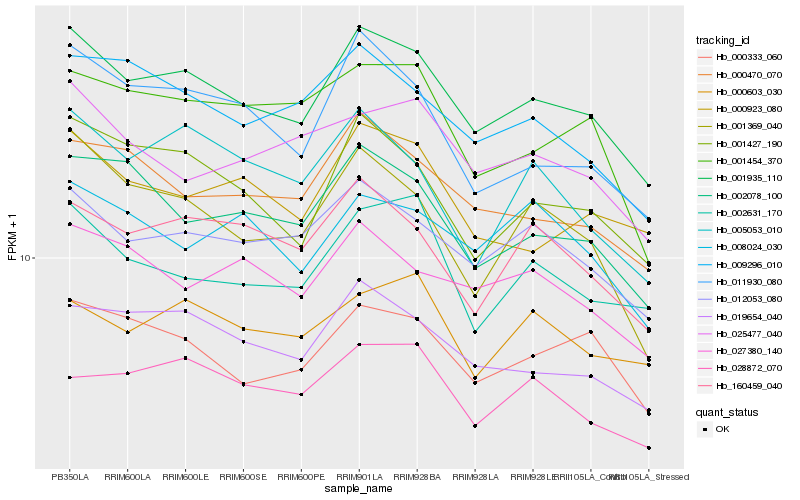
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001935_110 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 2 |
Hb_012053_080 |
0.0546177147 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 3 |
Hb_019654_040 |
0.0573921651 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001427_190 |
0.0601969194 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 5 |
Hb_000333_060 |
0.0626034095 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 6 |
Hb_000470_070 |
0.0637617917 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 7 |
Hb_002078_100 |
0.0656085458 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 8 |
Hb_009296_010 |
0.0689545384 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 9 |
Hb_160459_040 |
0.0693624433 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 10 |
Hb_011930_080 |
0.0710423412 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 11 |
Hb_027380_140 |
0.0733210757 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000923_080 |
0.0740897032 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 13 |
Hb_002631_170 |
0.0743123935 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 14 |
Hb_001369_040 |
0.0754081828 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_028872_070 |
0.0777197429 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001454_370 |
0.0788975308 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 17 |
Hb_025477_040 |
0.0792137251 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 18 |
Hb_008024_030 |
0.0806669491 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 19 |
Hb_000603_030 |
0.0809157922 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 20 |
Hb_005053_010 |
0.081367074 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |