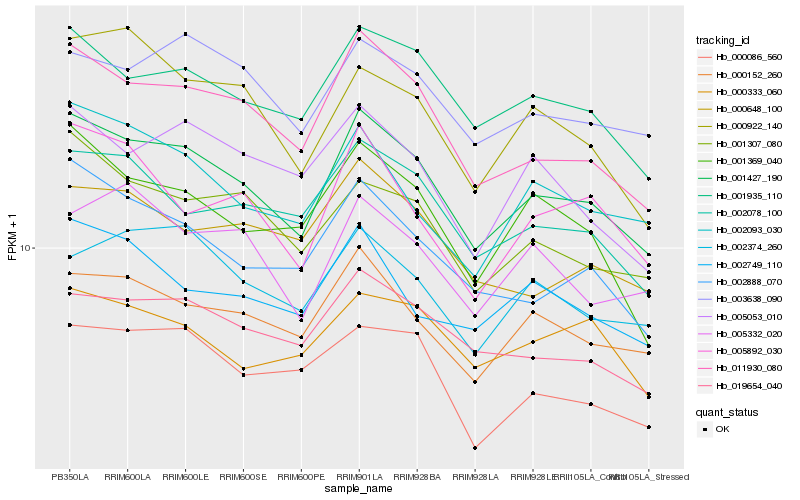
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_190 |
0.0 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 2 |
Hb_011930_080 |
0.051266689 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 3 |
Hb_001935_110 |
0.0601969194 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 4 |
Hb_000152_260 |
0.065158798 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 5 |
Hb_019654_040 |
0.0669390794 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002078_100 |
0.0703366708 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 7 |
Hb_002888_070 |
0.0723238367 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 8 |
Hb_001307_080 |
0.073850895 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 9 |
Hb_003638_090 |
0.0745745284 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 10 |
Hb_001369_040 |
0.078007415 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005053_010 |
0.0794782372 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 12 |
Hb_002093_030 |
0.0794833228 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 13 |
Hb_002374_260 |
0.0799873424 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 14 |
Hb_002749_110 |
0.0809531199 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000922_140 |
0.0816327809 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 16 |
Hb_005892_030 |
0.0823495613 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 17 |
Hb_000086_560 |
0.0828514912 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000648_100 |
0.0830914675 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 19 |
Hb_000333_060 |
0.0832551769 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 20 |
Hb_005332_020 |
0.0837190543 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |