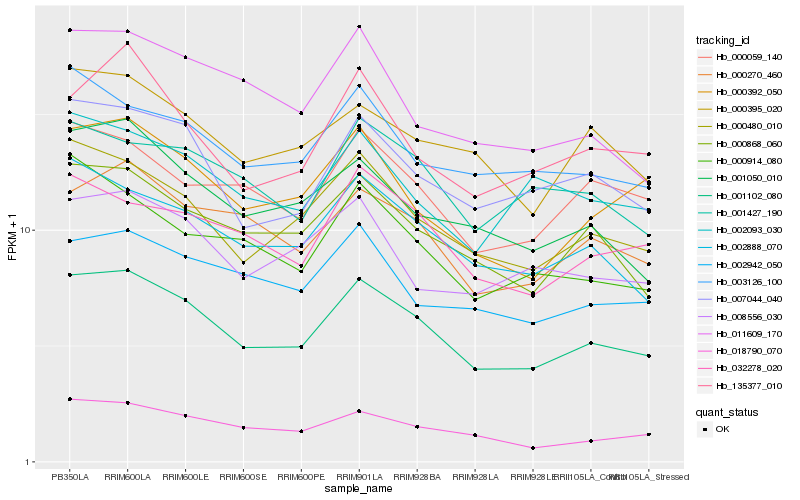
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_080 |
0.0 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 2 |
Hb_000480_010 |
0.0628754544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002888_070 |
0.0693916265 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 4 |
Hb_007044_040 |
0.0697057161 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 5 |
Hb_000868_060 |
0.0705987887 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_000270_460 |
0.0740153575 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 7 |
Hb_018790_070 |
0.0744104868 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 8 |
Hb_032278_020 |
0.0807408714 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001050_010 |
0.0821693561 |
- |
- |
Advillin [Gossypium arboreum] |
| 10 |
Hb_000059_140 |
0.0822212341 |
- |
- |
rnase h, putative [Ricinus communis] |
| 11 |
Hb_000395_020 |
0.0830130282 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 12 |
Hb_003126_100 |
0.0841571642 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 13 |
Hb_002942_050 |
0.0844605513 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000392_050 |
0.0852617055 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001427_190 |
0.0872419108 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 16 |
Hb_000914_080 |
0.0883292695 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_008556_030 |
0.0899114518 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011609_170 |
0.0907674164 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 19 |
Hb_135377_010 |
0.0908745048 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 20 |
Hb_002093_030 |
0.0915186409 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |