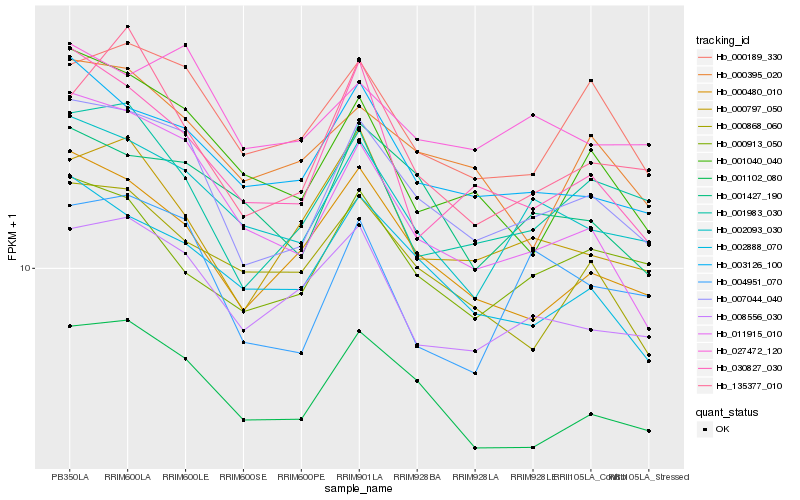
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 2 |
Hb_001102_080 |
0.0697057161 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_003126_100 |
0.079235678 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 4 |
Hb_002093_030 |
0.0792851 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 5 |
Hb_000480_010 |
0.0795308918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011915_010 |
0.0845464718 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 7 |
Hb_002888_070 |
0.0871444855 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 8 |
Hb_000189_330 |
0.0872163311 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 9 |
Hb_001983_030 |
0.0876031036 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 10 |
Hb_008556_030 |
0.0890779647 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001040_040 |
0.0924039827 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 12 |
Hb_001427_190 |
0.0949072186 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 13 |
Hb_000395_020 |
0.0952774756 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 14 |
Hb_000913_050 |
0.0957843235 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 15 |
Hb_027472_120 |
0.0957955356 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_030827_030 |
0.0964912901 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 17 |
Hb_004951_070 |
0.099012353 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 18 |
Hb_000797_050 |
0.0993501972 |
- |
- |
PREDICTED: uncharacterized protein LOC105645916 [Jatropha curcas] |
| 19 |
Hb_000868_060 |
0.101002263 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 20 |
Hb_135377_010 |
0.1012407885 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |