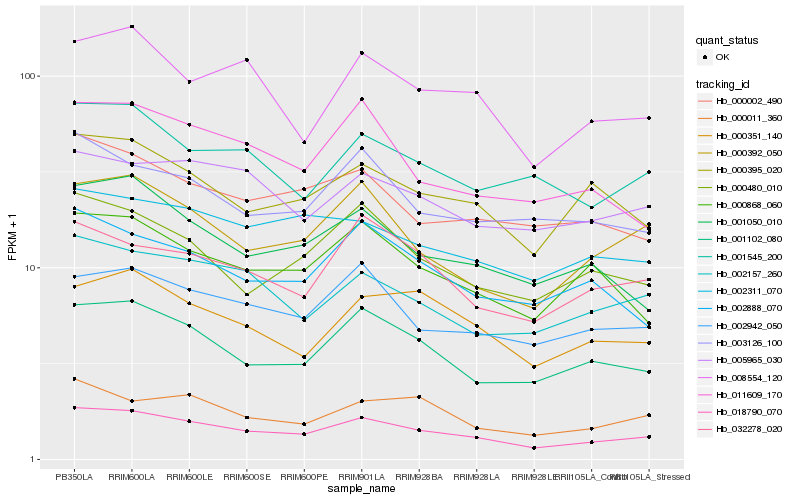
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018790_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 2 |
Hb_001102_080 |
0.0744104868 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_002942_050 |
0.0842411085 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000392_050 |
0.08452757 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002157_260 |
0.0868730637 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 6 |
Hb_000395_020 |
0.0882860957 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 7 |
Hb_000480_010 |
0.0884366287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001050_010 |
0.0885161947 |
- |
- |
Advillin [Gossypium arboreum] |
| 9 |
Hb_000868_060 |
0.0894584331 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 10 |
Hb_000011_360 |
0.0914894933 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000002_490 |
0.0917308248 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 12 |
Hb_002311_070 |
0.0919216623 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 13 |
Hb_008554_120 |
0.0919570973 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 14 |
Hb_032278_020 |
0.0921071134 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001545_200 |
0.0936646944 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 16 |
Hb_011609_170 |
0.0938800701 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 17 |
Hb_002888_070 |
0.0939039289 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 18 |
Hb_003126_100 |
0.0951504431 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 19 |
Hb_005965_030 |
0.0953064402 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 20 |
Hb_000351_140 |
0.095683303 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 9A-like isoform X3 [Pyrus x bretschneideri] |