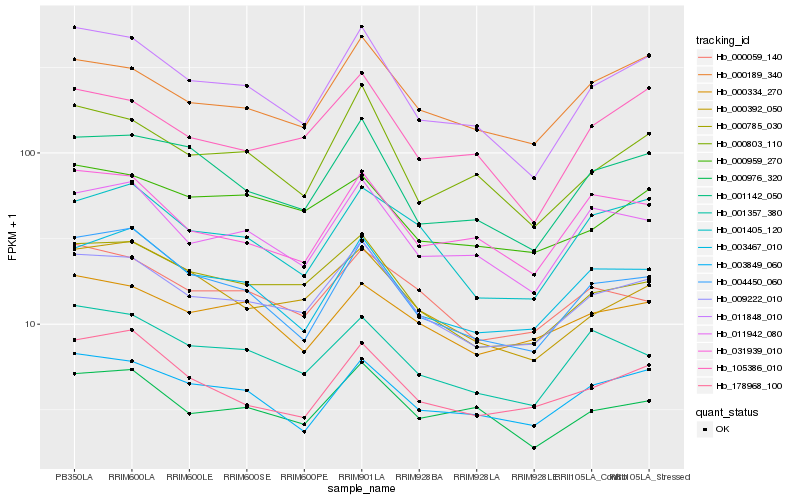
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_010 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_000785_030 |
0.055139497 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 3 |
Hb_011848_010 |
0.0679167164 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 4 |
Hb_000189_340 |
0.0687151055 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 5 |
Hb_000059_140 |
0.070665113 |
- |
- |
rnase h, putative [Ricinus communis] |
| 6 |
Hb_011942_080 |
0.0731266603 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 7 |
Hb_003467_010 |
0.0737869248 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_001142_050 |
0.0763909789 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 9 |
Hb_000392_050 |
0.0778028739 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003849_060 |
0.0781068519 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001357_380 |
0.0785837622 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000334_270 |
0.0828270249 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004450_060 |
0.0831695787 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000976_320 |
0.0835726429 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 15 |
Hb_000959_270 |
0.0841050982 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 16 |
Hb_001405_120 |
0.0842874376 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 17 |
Hb_178968_100 |
0.0843154437 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 18 |
Hb_000803_110 |
0.084631496 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 19 |
Hb_105386_010 |
0.084670605 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 20 |
Hb_031939_010 |
0.0848713956 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |