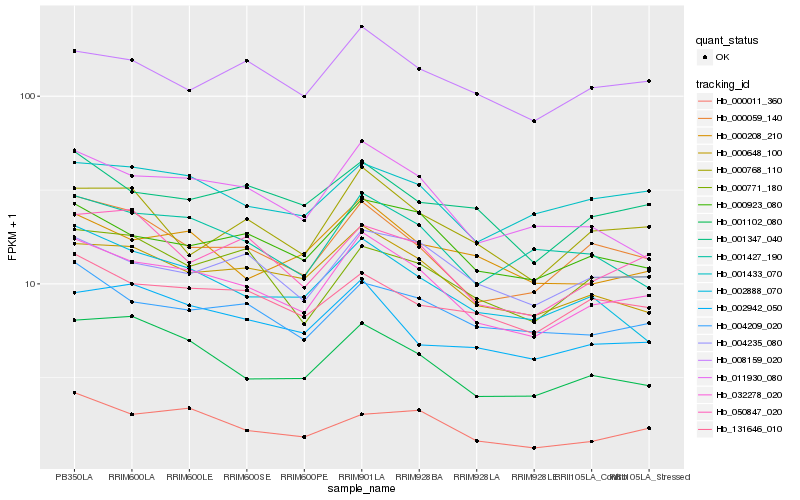
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032278_020 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000059_140 |
0.065584927 |
- |
- |
rnase h, putative [Ricinus communis] |
| 3 |
Hb_000648_100 |
0.0713700261 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 4 |
Hb_000923_080 |
0.0723998822 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 5 |
Hb_011930_080 |
0.077001304 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 6 |
Hb_001102_080 |
0.0807408714 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 7 |
Hb_004209_020 |
0.081332332 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001427_190 |
0.0837286129 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 9 |
Hb_004235_080 |
0.0855547218 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 10 |
Hb_001433_070 |
0.0863006251 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_008159_020 |
0.0868044901 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 12 |
Hb_000011_360 |
0.0869360635 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_002888_070 |
0.0869853124 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 14 |
Hb_050847_020 |
0.0880878864 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000768_110 |
0.0881426656 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001347_040 |
0.0881960899 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 17 |
Hb_131646_010 |
0.0887831566 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000208_210 |
0.0899479211 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 19 |
Hb_002942_050 |
0.0899973137 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000771_180 |
0.0909998678 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |