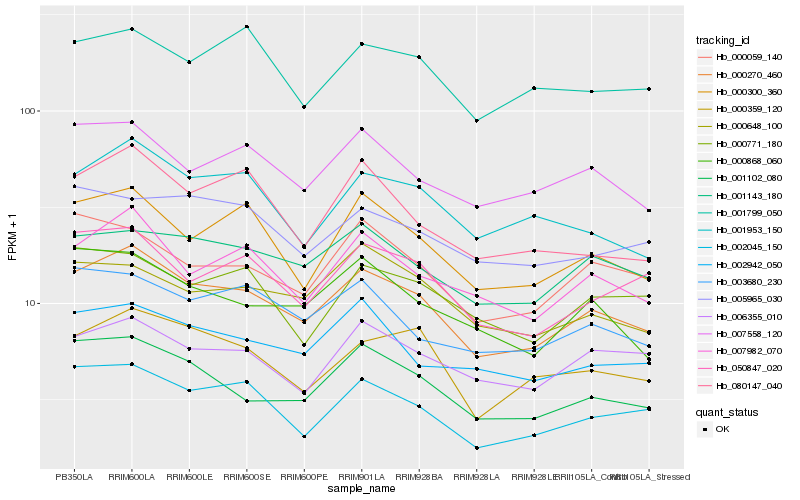
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_460 |
0.0 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 2 |
Hb_007982_070 |
0.0689937292 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 3 |
Hb_001102_080 |
0.0740153575 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 4 |
Hb_000300_360 |
0.0744489797 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 5 |
Hb_001143_180 |
0.0748343711 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 6 |
Hb_002045_150 |
0.0753671301 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 7 |
Hb_007558_120 |
0.0774719386 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 8 |
Hb_000359_120 |
0.0798955544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001953_150 |
0.0811828467 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 10 |
Hb_050847_020 |
0.0812960683 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_002942_050 |
0.0818583731 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003680_230 |
0.0839798944 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 13 |
Hb_000059_140 |
0.0842016378 |
- |
- |
rnase h, putative [Ricinus communis] |
| 14 |
Hb_000868_060 |
0.0848833407 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_006355_010 |
0.0848856068 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 16 |
Hb_000648_100 |
0.0884651741 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 17 |
Hb_080147_040 |
0.0901478776 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 18 |
Hb_001799_050 |
0.090151168 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 19 |
Hb_000771_180 |
0.0910613435 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 20 |
Hb_005965_030 |
0.0914235977 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |