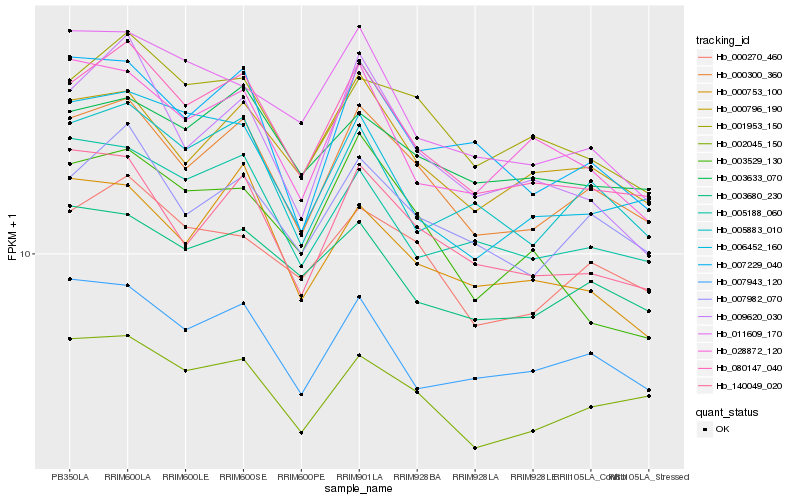
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 2 |
Hb_000300_360 |
0.0645797902 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 3 |
Hb_009620_030 |
0.076489822 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 4 |
Hb_001953_150 |
0.0780886064 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 5 |
Hb_006452_160 |
0.0813543665 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 6 |
Hb_005188_060 |
0.0828919066 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_140049_020 |
0.0837520976 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 8 |
Hb_007982_070 |
0.0838995667 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 9 |
Hb_005883_010 |
0.0848702051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000796_190 |
0.0850914335 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 11 |
Hb_011609_170 |
0.0868681336 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 12 |
Hb_007943_120 |
0.0885255513 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 13 |
Hb_007229_040 |
0.0895027397 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 14 |
Hb_028872_120 |
0.0901368202 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 15 |
Hb_000753_100 |
0.0901399521 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000270_460 |
0.0901478776 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 17 |
Hb_003633_070 |
0.0904555523 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 18 |
Hb_003529_130 |
0.0906784156 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_003680_230 |
0.0908090947 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 20 |
Hb_002045_150 |
0.0922566714 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |