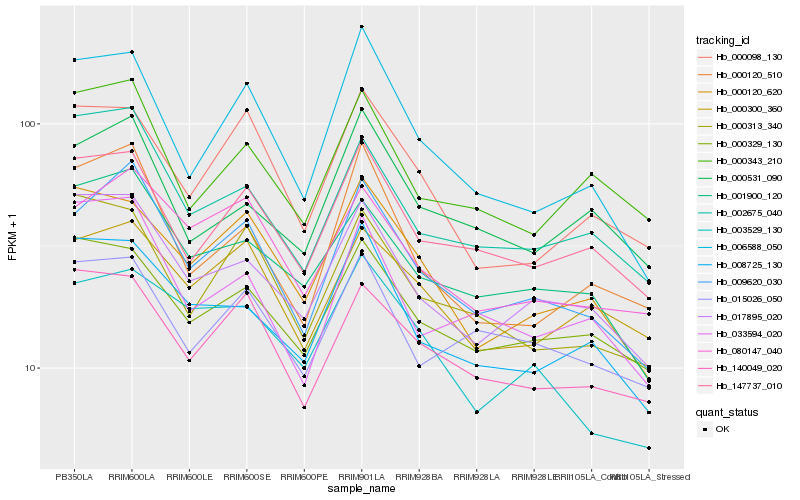
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009620_030 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 2 |
Hb_080147_040 |
0.076489822 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 3 |
Hb_001900_120 |
0.0841824458 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 4 |
Hb_017895_020 |
0.0854718039 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 5 |
Hb_147737_010 |
0.0867492854 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002675_040 |
0.0881251266 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 7 |
Hb_006588_050 |
0.0886562397 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 8 |
Hb_000120_510 |
0.0920896188 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_140049_020 |
0.0927673286 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 10 |
Hb_008725_130 |
0.0961703993 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 11 |
Hb_000300_360 |
0.0965032015 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 12 |
Hb_003529_130 |
0.0966107233 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000120_620 |
0.0974610243 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 14 |
Hb_033594_020 |
0.0976403544 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 15 |
Hb_000329_130 |
0.099887066 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 16 |
Hb_015026_050 |
0.1000628604 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000313_340 |
0.100757308 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 18 |
Hb_000531_090 |
0.1024427889 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 19 |
Hb_000098_130 |
0.1028611276 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 20 |
Hb_000343_210 |
0.1034303933 |
- |
- |
ran-binding protein, putative [Ricinus communis] |