| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
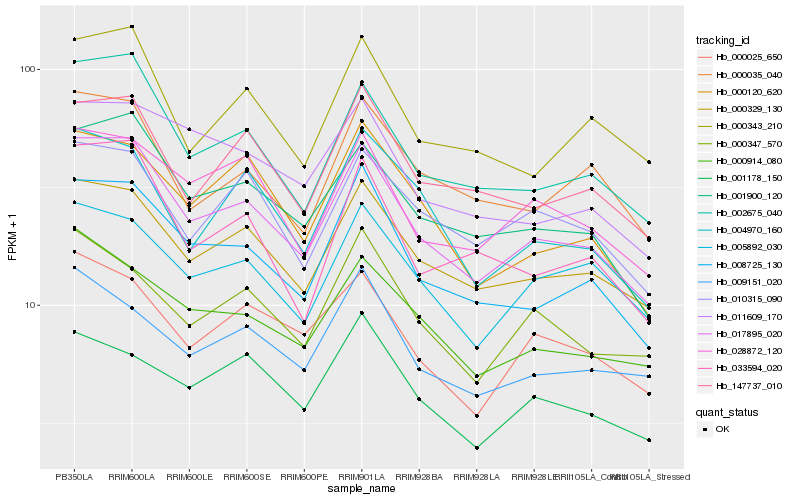
Hb_017895_020 |
0.0 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 2 |
Hb_002675_040 |
0.052133169 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000329_130 |
0.0531297769 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 4 |
Hb_001900_120 |
0.0538848603 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 5 |
Hb_008725_130 |
0.0567583674 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 6 |
Hb_028872_120 |
0.0719247505 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 7 |
Hb_000914_080 |
0.0741723062 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_147737_010 |
0.0753740692 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000025_650 |
0.0760822662 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 10 |
Hb_000120_620 |
0.0780031883 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 11 |
Hb_005892_030 |
0.0789765715 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 12 |
Hb_033594_020 |
0.0793447605 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 13 |
Hb_000343_210 |
0.079831861 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 14 |
Hb_010315_090 |
0.0799100528 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 15 |
Hb_000347_570 |
0.0811684224 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 16 |
Hb_004970_160 |
0.0812021303 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_011609_170 |
0.0821341746 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 18 |
Hb_000035_040 |
0.0824192056 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 19 |
Hb_001178_150 |
0.0847100982 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 20 |
Hb_009151_020 |
0.0849097465 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |