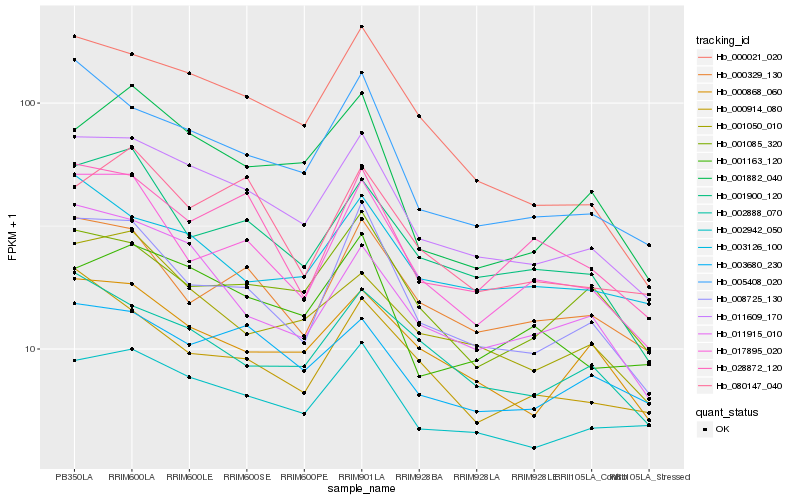
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_170 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 2 |
Hb_008725_130 |
0.0688670271 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 3 |
Hb_005408_020 |
0.0692568218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002942_050 |
0.0740645982 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000868_060 |
0.0769457378 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_001050_010 |
0.0775114929 |
- |
- |
Advillin [Gossypium arboreum] |
| 7 |
Hb_001163_120 |
0.0775560058 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_001900_120 |
0.0787611287 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 9 |
Hb_003680_230 |
0.0806671552 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 10 |
Hb_011915_010 |
0.0819737236 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 11 |
Hb_000021_020 |
0.0820228153 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 12 |
Hb_017895_020 |
0.0821341746 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 13 |
Hb_002888_070 |
0.0829755803 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 14 |
Hb_000329_130 |
0.0867593738 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 15 |
Hb_080147_040 |
0.0868681336 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 16 |
Hb_001882_040 |
0.0879494128 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 17 |
Hb_003126_100 |
0.0891915863 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 18 |
Hb_001085_320 |
0.089372701 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_028872_120 |
0.090307312 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 20 |
Hb_000914_080 |
0.0903117231 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |