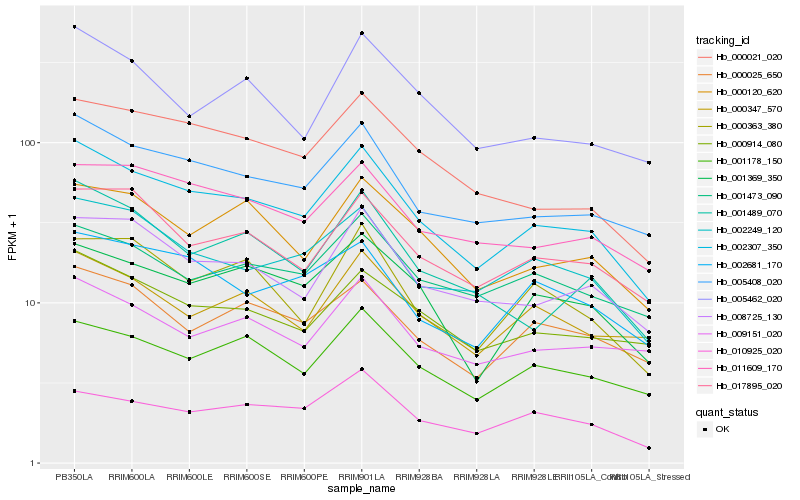
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_350 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_005408_020 |
0.063515994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002249_120 |
0.0797031329 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 4 |
Hb_008725_130 |
0.0838945345 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 5 |
Hb_017895_020 |
0.0852358061 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 6 |
Hb_000914_080 |
0.0867000735 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000021_020 |
0.0898810048 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 8 |
Hb_000120_620 |
0.0899417372 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 9 |
Hb_001369_350 |
0.0902780986 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 10 |
Hb_011609_170 |
0.091251411 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 11 |
Hb_001178_150 |
0.0925188605 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 12 |
Hb_000025_650 |
0.0929477084 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 13 |
Hb_002681_170 |
0.095364486 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 14 |
Hb_001489_070 |
0.0969370205 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 15 |
Hb_000363_380 |
0.0978397223 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 16 |
Hb_000347_570 |
0.0979307855 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 17 |
Hb_005462_020 |
0.0991720264 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 18 |
Hb_001473_090 |
0.1003823231 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 19 |
Hb_010925_020 |
0.1007561525 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 20 |
Hb_009151_020 |
0.1012086838 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |