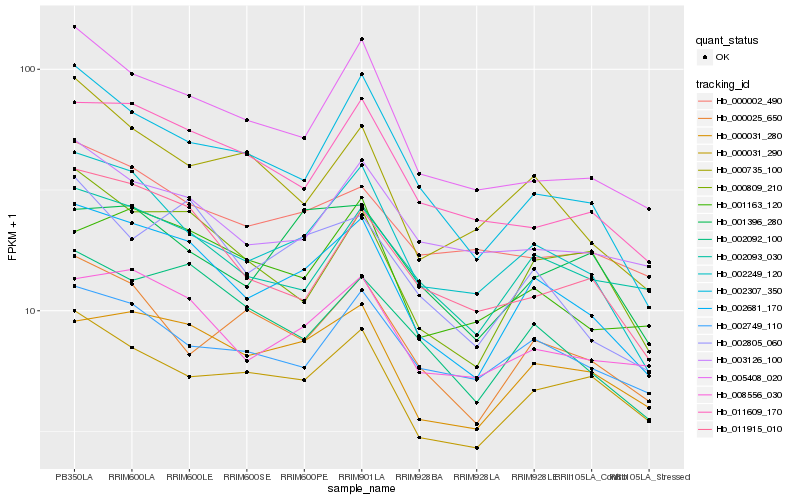
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002681_170 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 2 |
Hb_002249_120 |
0.0746546301 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000031_280 |
0.0818663794 |
- |
- |
- |
| 4 |
Hb_002092_100 |
0.086312491 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 5 |
Hb_008556_030 |
0.088310888 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002805_060 |
0.0899931049 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 7 |
Hb_000809_210 |
0.0929001266 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 8 |
Hb_002307_350 |
0.095364486 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_002093_030 |
0.0964719314 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 10 |
Hb_000031_290 |
0.0969829956 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 11 |
Hb_005408_020 |
0.0982330356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001163_120 |
0.1026957321 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_011609_170 |
0.1029307405 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 14 |
Hb_011915_010 |
0.1037782575 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 15 |
Hb_000025_650 |
0.105175679 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 16 |
Hb_000735_100 |
0.1081817733 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 17 |
Hb_001396_280 |
0.1082992372 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 18 |
Hb_002749_110 |
0.1093433479 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003126_100 |
0.1096007224 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 20 |
Hb_000002_490 |
0.1102175575 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |