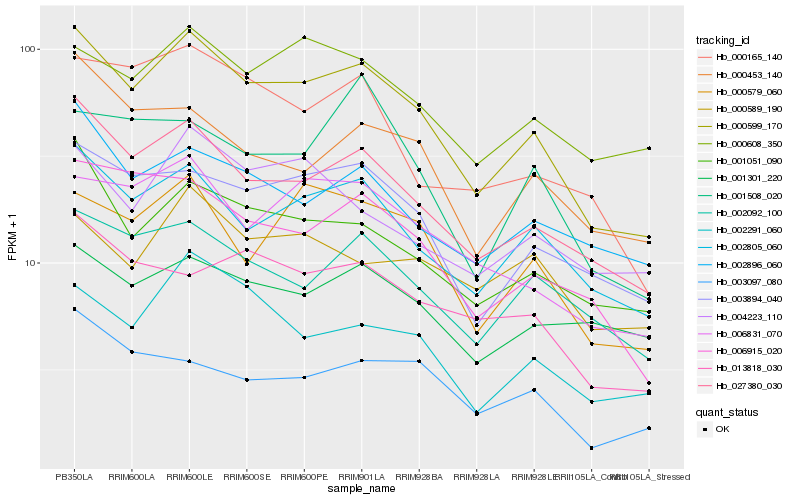
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_170 |
0.0 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 2 |
Hb_027380_030 |
0.0680463863 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002805_060 |
0.0808206238 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 4 |
Hb_003894_040 |
0.0929158391 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 5 |
Hb_001051_090 |
0.1002458514 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 6 |
Hb_006915_020 |
0.1052288409 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 7 |
Hb_002092_100 |
0.1059980847 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 8 |
Hb_006831_070 |
0.1062383495 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000579_060 |
0.1087366688 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002291_060 |
0.1112134794 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_002896_060 |
0.1133156172 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 12 |
Hb_000608_350 |
0.113548539 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 13 |
Hb_000165_140 |
0.1165648543 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 14 |
Hb_000453_140 |
0.1187883317 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 15 |
Hb_013818_030 |
0.1212651081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004223_110 |
0.122234289 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 17 |
Hb_003097_080 |
0.1243409958 |
- |
- |
hypothetical protein JCGZ_24481 [Jatropha curcas] |
| 18 |
Hb_000589_190 |
0.1287953488 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001301_220 |
0.1288423609 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 20 |
Hb_001508_020 |
0.1330855248 |
- |
- |
myosin vIII, putative [Ricinus communis] |