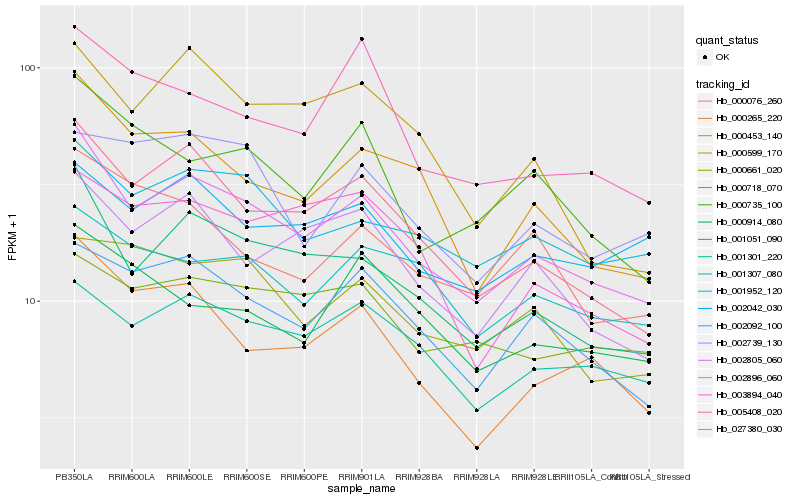
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002896_060 |
0.0 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 2 |
Hb_001051_090 |
0.0628655337 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 3 |
Hb_027380_030 |
0.0714348837 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002092_100 |
0.0869658928 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 5 |
Hb_000453_140 |
0.089516711 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 6 |
Hb_001952_120 |
0.0911667538 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002805_060 |
0.0926823073 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 8 |
Hb_001301_220 |
0.0928365643 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 9 |
Hb_000265_220 |
0.0996955025 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 10 |
Hb_000735_100 |
0.1037195478 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 11 |
Hb_002042_030 |
0.107519397 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 12 |
Hb_000718_070 |
0.1079415221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001307_080 |
0.1082537982 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 14 |
Hb_005408_020 |
0.1085726075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000076_260 |
0.1095907453 |
- |
- |
hypothetical protein GUITHDRAFT_149979 [Guillardia theta CCMP2712] |
| 16 |
Hb_000661_020 |
0.1109840023 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 17 |
Hb_000599_170 |
0.1133156172 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_003894_040 |
0.1148832249 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 19 |
Hb_000914_080 |
0.1153605307 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_002739_130 |
0.1154176802 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |