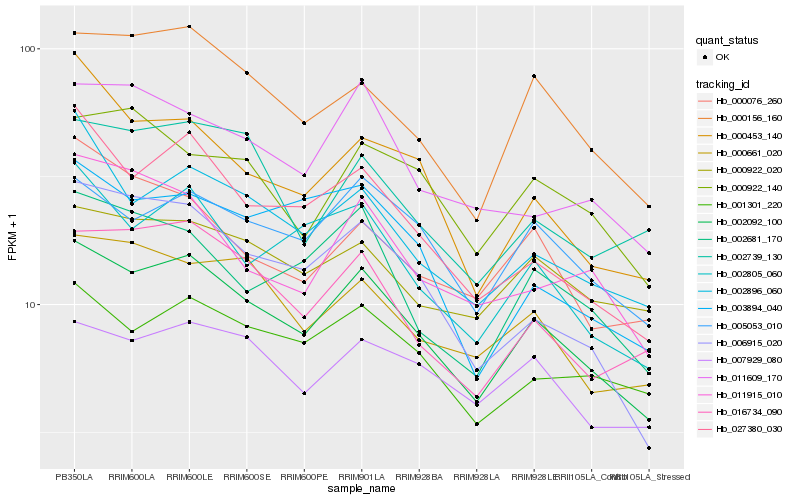
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002092_100 |
0.0 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 2 |
Hb_000156_160 |
0.0716702443 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 3 |
Hb_005053_010 |
0.0795145886 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 4 |
Hb_002805_060 |
0.0801579285 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 5 |
Hb_027380_030 |
0.0831401921 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_016734_090 |
0.0847390054 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 7 |
Hb_002681_170 |
0.086312491 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 8 |
Hb_002896_060 |
0.0869658928 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 9 |
Hb_001301_220 |
0.0870149843 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 10 |
Hb_000661_020 |
0.0879094234 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 11 |
Hb_006915_020 |
0.088117211 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 12 |
Hb_007929_080 |
0.0904205175 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 13 |
Hb_000922_020 |
0.093981735 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000453_140 |
0.0947227353 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 15 |
Hb_000922_140 |
0.0953735628 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 16 |
Hb_011609_170 |
0.0955667659 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 17 |
Hb_011915_010 |
0.0998996933 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 18 |
Hb_003894_040 |
0.0999542525 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 19 |
Hb_000076_260 |
0.1003337758 |
- |
- |
hypothetical protein GUITHDRAFT_149979 [Guillardia theta CCMP2712] |
| 20 |
Hb_002739_130 |
0.1004126816 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |