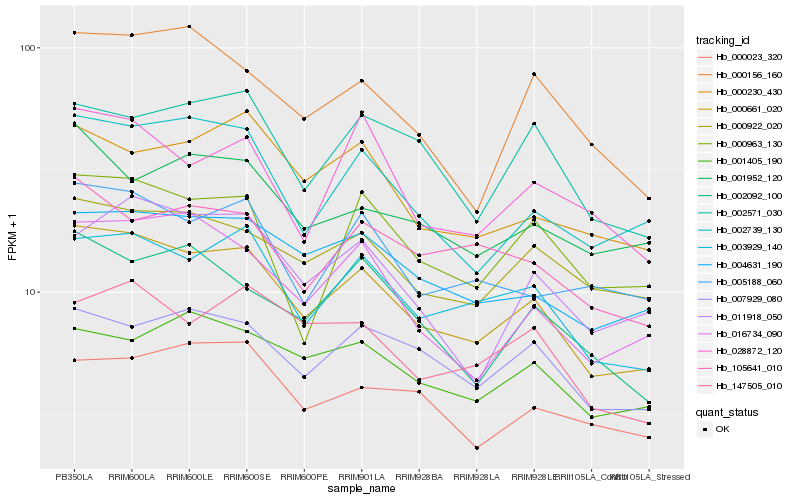
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_020 |
0.0 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 2 |
Hb_003929_140 |
0.0646130366 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_004631_190 |
0.0747685552 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 4 |
Hb_002739_130 |
0.0769563564 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 5 |
Hb_016734_090 |
0.0821170386 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000922_020 |
0.0835473247 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_007929_080 |
0.0849057705 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_005188_060 |
0.0869421436 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 9 |
Hb_002092_100 |
0.0879094234 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 10 |
Hb_147505_010 |
0.0898159532 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 11 |
Hb_000230_430 |
0.0898898327 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000156_160 |
0.091386804 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 13 |
Hb_001405_190 |
0.0938275964 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_105641_010 |
0.0950108165 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_001952_120 |
0.0951698819 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002571_030 |
0.0954352369 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 17 |
Hb_011918_050 |
0.0956394575 |
- |
- |
- |
| 18 |
Hb_028872_120 |
0.0966440342 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 19 |
Hb_000963_130 |
0.0973304057 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000023_320 |
0.098622291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |