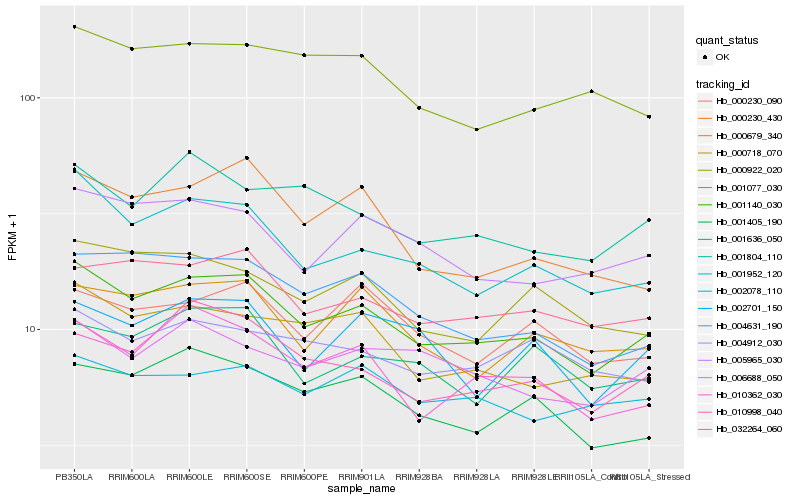
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 2 |
Hb_001952_120 |
0.057552329 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_010998_040 |
0.062716695 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 4 |
Hb_032264_060 |
0.0668190619 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 5 |
Hb_004631_190 |
0.0699857195 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 6 |
Hb_005965_030 |
0.0703095098 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 7 |
Hb_001405_190 |
0.0712786685 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000679_340 |
0.0738033457 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000718_070 |
0.0756336374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010362_030 |
0.0776107103 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_001804_110 |
0.0778357104 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 12 |
Hb_000922_020 |
0.0783571568 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_006688_050 |
0.0787234548 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000230_090 |
0.0805310851 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001636_050 |
0.080545495 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 16 |
Hb_002078_110 |
0.0808980233 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 17 |
Hb_001077_030 |
0.0815605209 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000230_430 |
0.0822015023 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_002701_150 |
0.0830150627 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004912_030 |
0.0839840328 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |