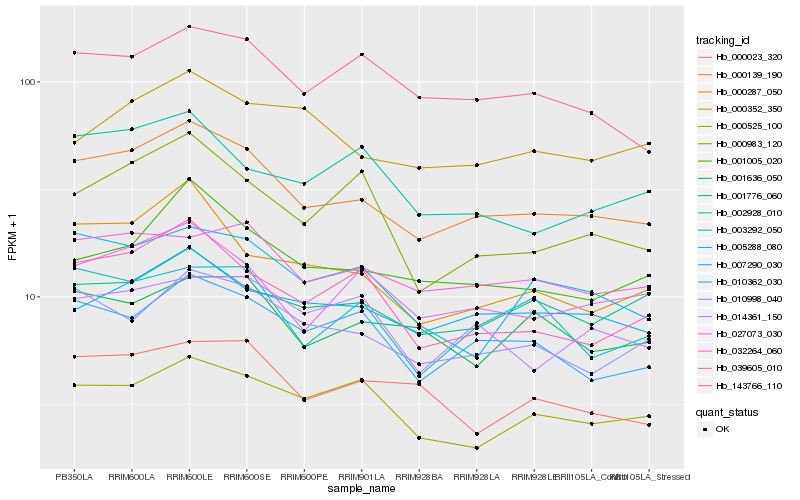
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_190 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_027073_030 |
0.0741547857 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 3 |
Hb_010998_040 |
0.0763425878 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 4 |
Hb_039605_010 |
0.0779254529 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 5 |
Hb_032264_060 |
0.0785943761 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 6 |
Hb_010362_030 |
0.0799051694 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_000287_050 |
0.0799486247 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 8 |
Hb_001005_020 |
0.0800580311 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 9 |
Hb_007290_030 |
0.0847875855 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 10 |
Hb_143766_110 |
0.0853044498 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 11 |
Hb_005288_080 |
0.0893305838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001776_060 |
0.0899882937 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 13 |
Hb_000983_120 |
0.0916016106 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000525_100 |
0.09171643 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 15 |
Hb_014361_150 |
0.0922668906 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 16 |
Hb_003292_050 |
0.0924199261 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 17 |
Hb_000023_320 |
0.093087668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001636_050 |
0.0935060303 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 19 |
Hb_000352_350 |
0.0938414484 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002928_010 |
0.0939578593 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |