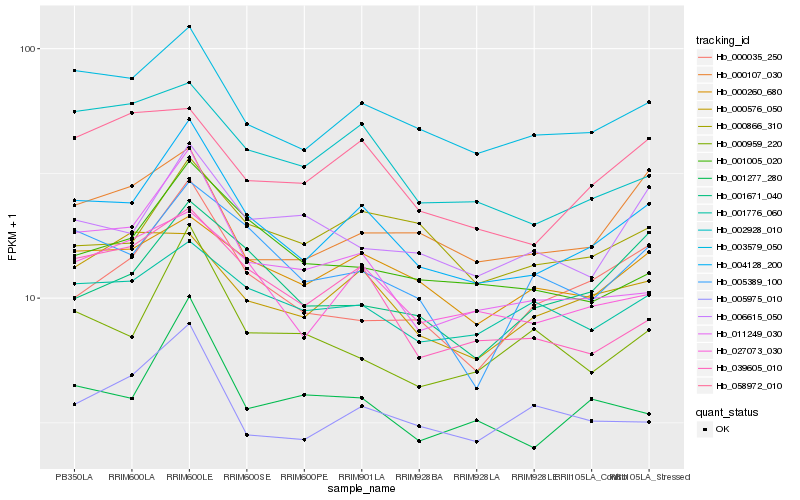
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 2 |
Hb_003579_050 |
0.0720900208 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 3 |
Hb_011249_030 |
0.0832299917 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 4 |
Hb_027073_030 |
0.0844834508 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 5 |
Hb_001005_020 |
0.0957521528 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 6 |
Hb_001277_280 |
0.0958023104 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 7 |
Hb_039605_010 |
0.0980875369 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_680 |
0.0989353401 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 9 |
Hb_000035_250 |
0.0994775609 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 10 |
Hb_002928_010 |
0.1003118094 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 11 |
Hb_005389_100 |
0.1005461014 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 12 |
Hb_058972_010 |
0.1023176244 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001776_060 |
0.102824422 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 14 |
Hb_005975_010 |
0.1028866953 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000866_310 |
0.1033511605 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 16 |
Hb_006615_050 |
0.1043492769 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 17 |
Hb_001671_040 |
0.1043881292 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000107_030 |
0.1056975254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000959_220 |
0.1061600647 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000576_050 |
0.106303433 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |