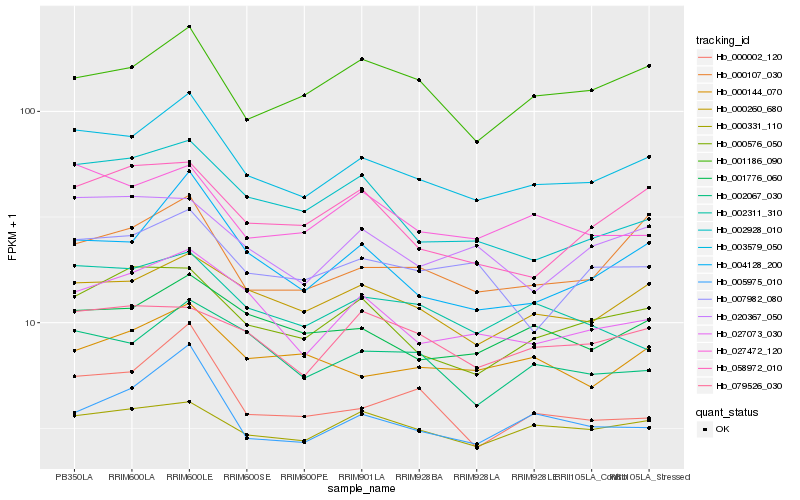
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003579_050 |
0.0 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 2 |
Hb_000260_680 |
0.0695891137 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 3 |
Hb_004128_200 |
0.0720900208 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 4 |
Hb_027073_030 |
0.073671087 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 5 |
Hb_001776_060 |
0.0749730891 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 6 |
Hb_000107_030 |
0.0774789614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_027472_120 |
0.0775348077 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_005975_010 |
0.0789672318 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_002067_030 |
0.0799011868 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000576_050 |
0.0803931214 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 11 |
Hb_001186_090 |
0.0832393517 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 12 |
Hb_000002_120 |
0.0841402501 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 13 |
Hb_007982_080 |
0.0864521662 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 14 |
Hb_020367_050 |
0.0871333324 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_002928_010 |
0.0878622435 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 16 |
Hb_000331_110 |
0.0879805582 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 17 |
Hb_002311_310 |
0.0882331589 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 18 |
Hb_000144_070 |
0.0887959662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_079526_030 |
0.0889332428 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 20 |
Hb_058972_010 |
0.0908641019 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |