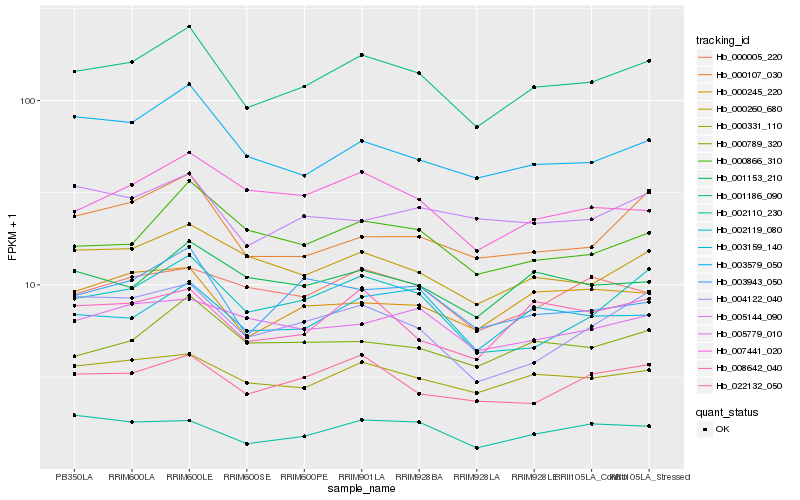
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001186_090 |
0.0 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 2 |
Hb_000331_110 |
0.0676286764 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 3 |
Hb_000260_680 |
0.0722129051 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 4 |
Hb_003943_050 |
0.0749498884 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 5 |
Hb_003159_140 |
0.0764307009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022132_050 |
0.077130972 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 7 |
Hb_002119_080 |
0.0777438394 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000789_320 |
0.0785340038 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000245_220 |
0.0786675477 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 10 |
Hb_001153_210 |
0.0808556982 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008642_040 |
0.0814957132 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000866_310 |
0.081516203 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 13 |
Hb_003579_050 |
0.0832393517 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 14 |
Hb_007441_020 |
0.0841072056 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 15 |
Hb_000005_220 |
0.0867594566 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 16 |
Hb_002110_230 |
0.0876927713 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_000107_030 |
0.0877057405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005144_090 |
0.0882304485 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 19 |
Hb_004122_040 |
0.0883646125 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_005779_010 |
0.0883709905 |
- |
- |
catalytic, putative [Ricinus communis] |