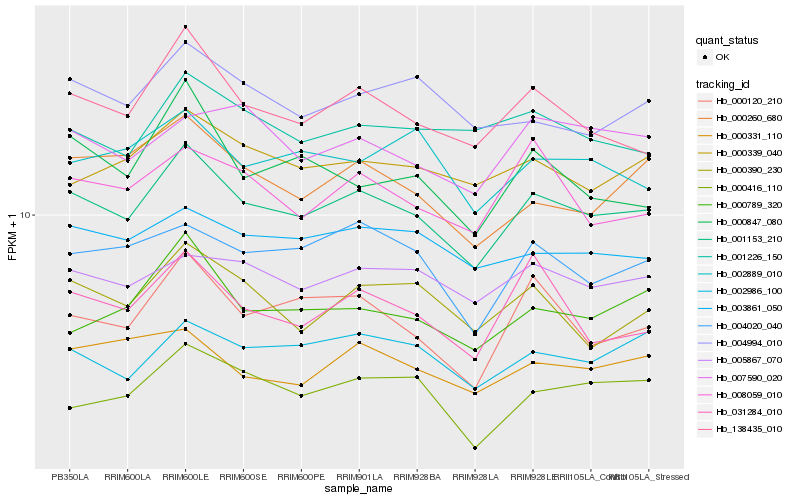
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 2 |
Hb_138435_010 |
0.0529780313 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 3 |
Hb_002986_100 |
0.0612319041 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 4 |
Hb_005867_070 |
0.0634544409 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_003861_050 |
0.0635194628 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 6 |
Hb_004020_040 |
0.0639539835 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000339_040 |
0.0659471626 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_000120_210 |
0.0662518049 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 9 |
Hb_000789_320 |
0.0663588058 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000260_680 |
0.0666002128 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 11 |
Hb_000390_230 |
0.0671942734 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_004994_010 |
0.0672359064 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_000331_110 |
0.0682507189 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 14 |
Hb_008059_010 |
0.0696596293 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001226_150 |
0.0698655946 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_031284_010 |
0.0699743076 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 17 |
Hb_000847_080 |
0.0711958041 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 18 |
Hb_007590_020 |
0.0712852207 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 19 |
Hb_000416_110 |
0.0714838391 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002889_010 |
0.0718505737 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |