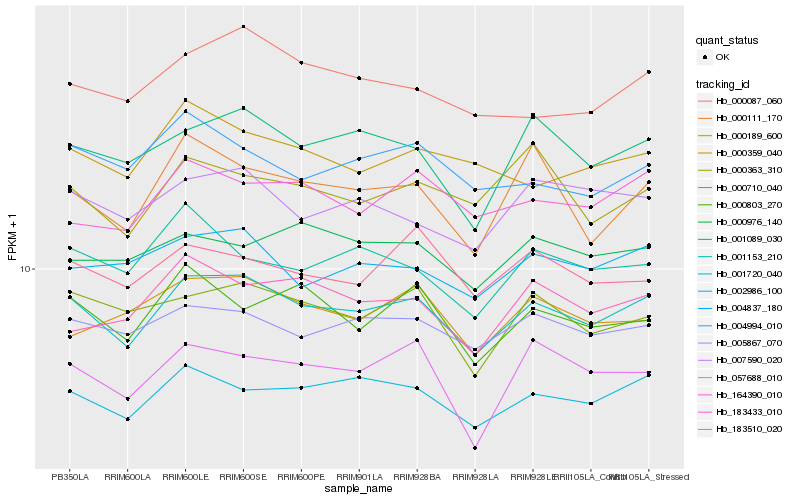
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001720_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002986_100 |
0.0518868794 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 3 |
Hb_005867_070 |
0.0588576694 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001089_030 |
0.0610647007 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 5 |
Hb_004837_180 |
0.0639989439 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000710_040 |
0.0641792928 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 7 |
Hb_007590_020 |
0.0665645607 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 8 |
Hb_164390_010 |
0.0676491991 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 9 |
Hb_000087_060 |
0.0686031111 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_000803_270 |
0.0698168428 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 11 |
Hb_000111_170 |
0.0702189562 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 12 |
Hb_183510_020 |
0.0709906989 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 13 |
Hb_000363_310 |
0.0709956688 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 14 |
Hb_000189_600 |
0.0710662574 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_183433_010 |
0.0710729716 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 16 |
Hb_004994_010 |
0.0717542476 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 17 |
Hb_000976_140 |
0.0726848315 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001153_210 |
0.0729265491 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000359_040 |
0.0731380001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_057688_010 |
0.0733248487 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |