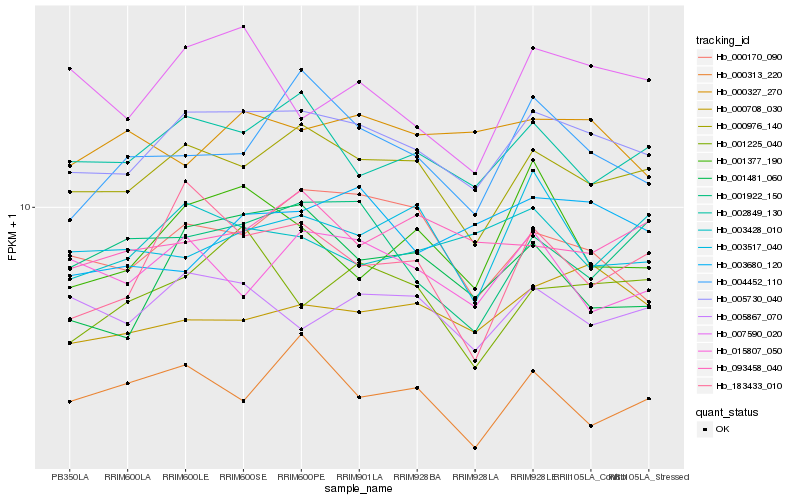
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005730_040 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 2 |
Hb_000976_140 |
0.0347572996 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002849_130 |
0.0483417487 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 4 |
Hb_005867_070 |
0.0515346819 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000708_030 |
0.0559825597 |
- |
- |
PREDICTED: uncharacterized protein LOC105650742 [Jatropha curcas] |
| 6 |
Hb_000170_090 |
0.0565072701 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 7 |
Hb_001225_040 |
0.0591048195 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 8 |
Hb_001481_060 |
0.0598947422 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004452_110 |
0.0600384702 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015807_050 |
0.0601998955 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 11 |
Hb_000313_220 |
0.0609041706 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000327_270 |
0.0610555316 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_093458_040 |
0.0611439445 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 14 |
Hb_003428_010 |
0.061318906 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 15 |
Hb_001922_150 |
0.0618519851 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_183433_010 |
0.0624303063 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 17 |
Hb_003517_040 |
0.0630223735 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007590_020 |
0.0637032202 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 19 |
Hb_001377_190 |
0.0645732304 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003680_120 |
0.0646371465 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL7 [Jatropha curcas] |