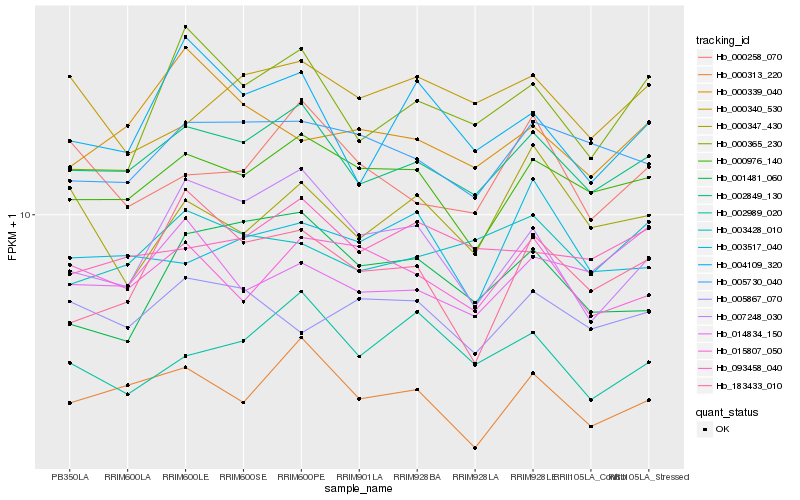
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_130 |
0.0 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 2 |
Hb_000313_220 |
0.0345223875 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000976_140 |
0.038434814 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005730_040 |
0.0483417487 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 5 |
Hb_093458_040 |
0.0497080707 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 6 |
Hb_003428_010 |
0.0525688406 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 7 |
Hb_015807_050 |
0.0544217542 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 8 |
Hb_002989_020 |
0.0546751358 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000258_070 |
0.0570037646 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 10 |
Hb_004109_320 |
0.0585974269 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 11 |
Hb_014834_150 |
0.0604271594 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000365_230 |
0.0606906014 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 13 |
Hb_007248_030 |
0.0612585676 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 14 |
Hb_000340_530 |
0.0621056011 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 15 |
Hb_005867_070 |
0.0641430094 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_001481_060 |
0.0653081049 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003517_040 |
0.0657142413 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 18 |
Hb_183433_010 |
0.0661019743 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 19 |
Hb_000347_430 |
0.0670483341 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 20 |
Hb_000339_040 |
0.0670968703 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |