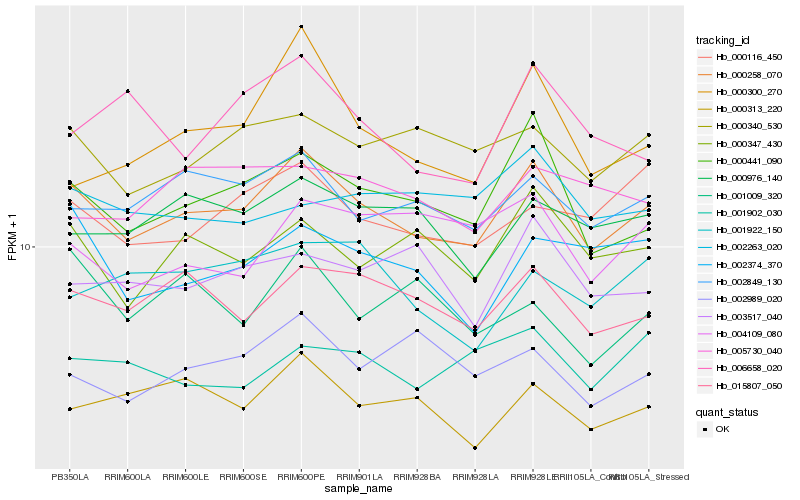
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 2 |
Hb_000441_090 |
0.0512305375 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 3 |
Hb_015807_050 |
0.0552153577 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 4 |
Hb_002849_130 |
0.0570037646 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 5 |
Hb_000340_530 |
0.060932849 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 6 |
Hb_000313_220 |
0.0615774455 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000347_430 |
0.0618165918 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 8 |
Hb_000116_450 |
0.0643643628 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000976_140 |
0.0649312325 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002374_370 |
0.0683039007 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_002989_020 |
0.0691905091 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001902_030 |
0.0707884665 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003517_040 |
0.0718078869 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005730_040 |
0.0737865893 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 15 |
Hb_001922_150 |
0.0738116907 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000300_270 |
0.0746887503 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_004109_080 |
0.074845747 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 18 |
Hb_002263_020 |
0.074950598 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 19 |
Hb_001009_320 |
0.0757821811 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 20 |
Hb_006658_020 |
0.0773014418 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |