| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
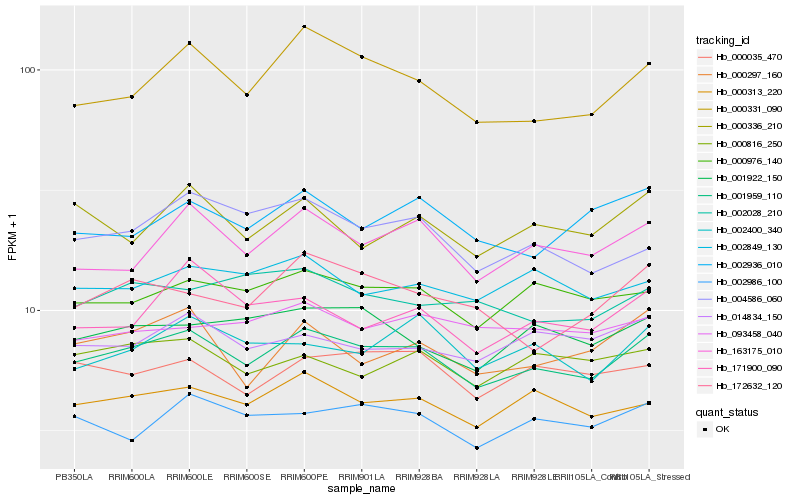
Hb_001959_110 |
0.0 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000313_220 |
0.0573220751 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000976_140 |
0.0622201152 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000331_090 |
0.0628708103 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 5 |
Hb_163175_010 |
0.0629477166 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 6 |
Hb_000035_470 |
0.0671842746 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001922_150 |
0.0681900878 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_002986_100 |
0.068607252 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 9 |
Hb_004586_060 |
0.0700598766 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 10 |
Hb_014834_150 |
0.0704068771 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000816_250 |
0.0708520571 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002849_130 |
0.0709266759 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 13 |
Hb_000297_160 |
0.0709674027 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 14 |
Hb_002400_340 |
0.0718707806 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 15 |
Hb_171900_090 |
0.0725087879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002936_010 |
0.0729490186 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 17 |
Hb_000336_210 |
0.0735356137 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 18 |
Hb_172632_120 |
0.0743005791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002028_210 |
0.0746691263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 20 |
Hb_093458_040 |
0.0749787922 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |