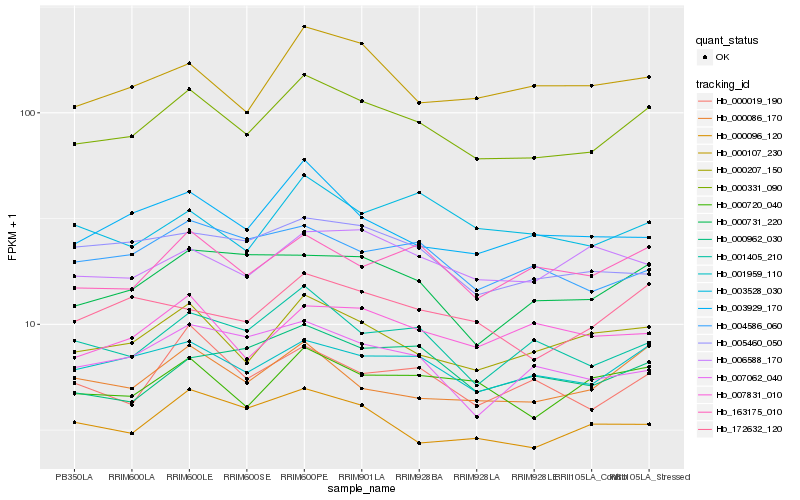
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_090 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 2 |
Hb_000207_150 |
0.0623332733 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 3 |
Hb_001959_110 |
0.0628708103 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 4 |
Hb_163175_010 |
0.0739569636 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 5 |
Hb_001405_210 |
0.0745624275 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 6 |
Hb_000720_040 |
0.0755179496 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 7 |
Hb_000107_230 |
0.0779255184 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_000086_170 |
0.0786825 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 9 |
Hb_172632_120 |
0.0788910054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000096_120 |
0.0794151908 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 11 |
Hb_000731_220 |
0.0795130509 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 12 |
Hb_003929_170 |
0.0807808978 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 13 |
Hb_000019_190 |
0.0816078631 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 14 |
Hb_004586_060 |
0.0848582722 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_007831_010 |
0.0857142911 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 16 |
Hb_000962_030 |
0.0858091755 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 17 |
Hb_005460_050 |
0.0864991679 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 18 |
Hb_003528_030 |
0.0867953621 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 19 |
Hb_007062_040 |
0.0868942313 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 20 |
Hb_006588_170 |
0.0870220395 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |