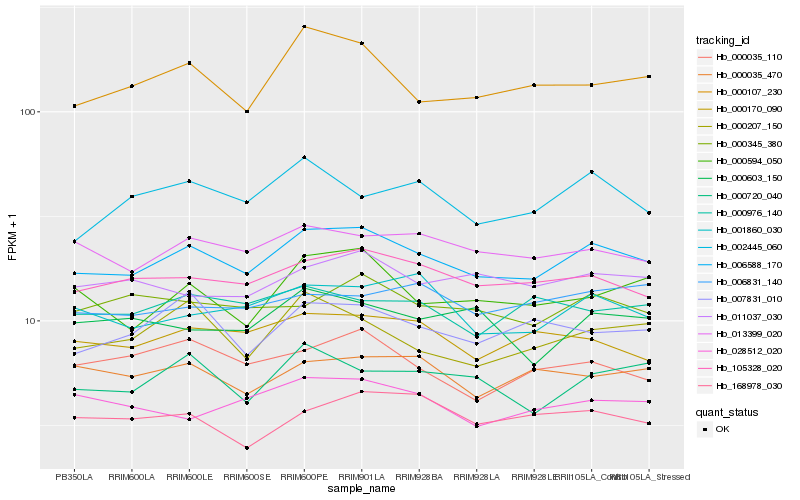
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_170 |
0.0 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 2 |
Hb_105328_020 |
0.056659463 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 3 |
Hb_013399_020 |
0.0637690547 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 4 |
Hb_000170_090 |
0.0694220737 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 5 |
Hb_000345_380 |
0.0707260559 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 6 |
Hb_001860_030 |
0.0715421116 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 7 |
Hb_000107_230 |
0.0723746057 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_011037_030 |
0.0726615901 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 9 |
Hb_007831_010 |
0.0738572208 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 10 |
Hb_000720_040 |
0.0738962655 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 11 |
Hb_000594_050 |
0.0748835495 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000035_470 |
0.0753456116 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000035_110 |
0.0755638746 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 14 |
Hb_000207_150 |
0.0756404464 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 15 |
Hb_002445_060 |
0.0760171681 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 16 |
Hb_028512_020 |
0.0762405668 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000976_140 |
0.0770557015 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000603_150 |
0.0787395485 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 19 |
Hb_006831_140 |
0.0788727196 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_168978_030 |
0.0794464998 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |