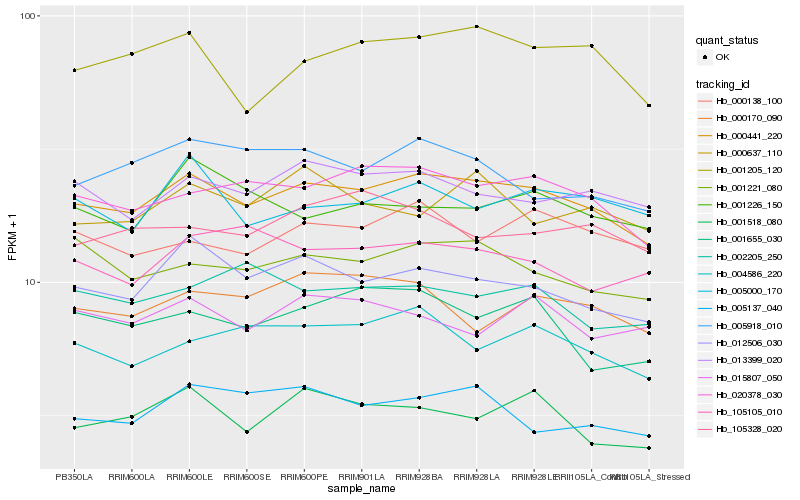
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_220 |
0.0 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_020378_030 |
0.0490381017 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 3 |
Hb_012506_030 |
0.0520882105 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 4 |
Hb_001221_080 |
0.0592097481 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_013399_020 |
0.0592957952 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 6 |
Hb_004586_220 |
0.0604709817 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 7 |
Hb_001205_120 |
0.0636696014 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 8 |
Hb_005918_010 |
0.0654786084 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 9 |
Hb_000138_100 |
0.0666172336 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 10 |
Hb_005137_040 |
0.0675131513 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 11 |
Hb_001518_080 |
0.0675464312 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 12 |
Hb_005000_170 |
0.0684629315 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 13 |
Hb_001655_030 |
0.0691867104 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_105105_010 |
0.0696938254 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 15 |
Hb_000170_090 |
0.0700909193 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 16 |
Hb_105328_020 |
0.0701060964 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 17 |
Hb_000637_110 |
0.0715839987 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 18 |
Hb_001226_150 |
0.0716998261 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_015807_050 |
0.0718845492 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 20 |
Hb_002205_250 |
0.0719616466 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |