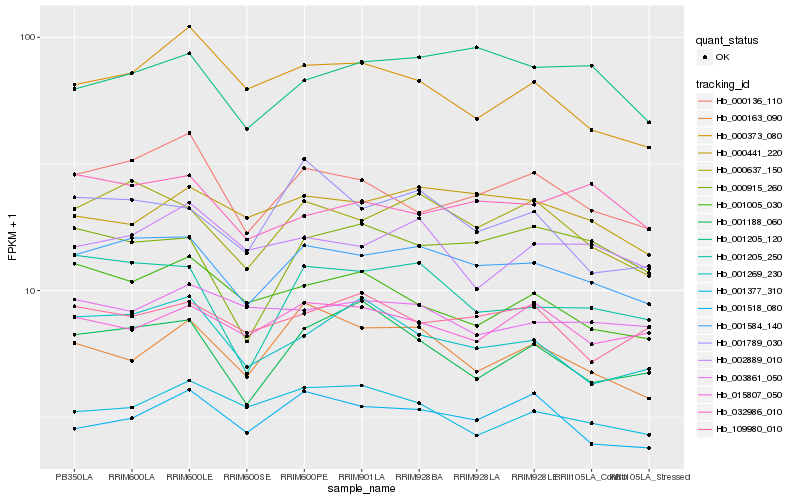
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 2 |
Hb_000637_150 |
0.0443903104 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 3 |
Hb_000136_110 |
0.0668173815 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 4 |
Hb_001205_250 |
0.069733717 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 5 |
Hb_000441_220 |
0.0725387687 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001205_120 |
0.0730812112 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 7 |
Hb_001518_080 |
0.0742836929 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 8 |
Hb_001269_230 |
0.0744708624 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 9 |
Hb_015807_050 |
0.0764316227 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 10 |
Hb_001789_030 |
0.0773482219 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 11 |
Hb_032986_010 |
0.0788673493 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 12 |
Hb_109980_010 |
0.0790107655 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000163_090 |
0.079843593 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 14 |
Hb_002889_010 |
0.0802359897 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 15 |
Hb_001005_030 |
0.0805902649 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_003861_050 |
0.0807751178 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 17 |
Hb_001188_060 |
0.0813784894 |
- |
- |
- |
| 18 |
Hb_000915_260 |
0.0818909052 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001377_310 |
0.0820089303 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 20 |
Hb_000373_080 |
0.0828855916 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |