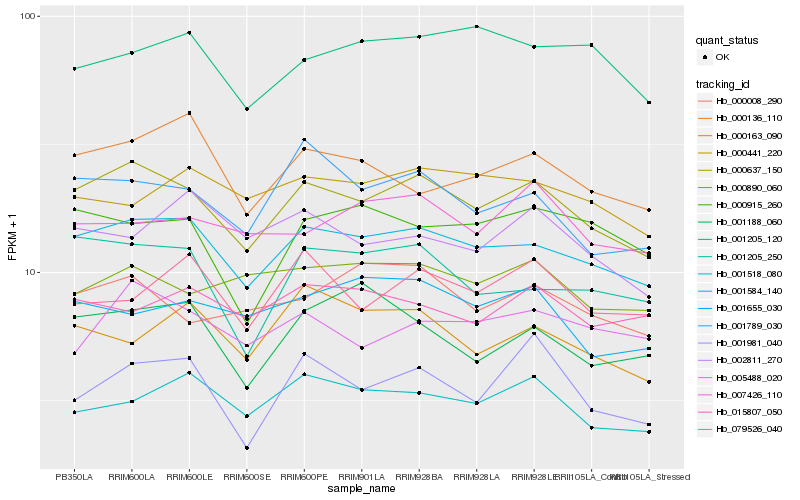
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_150 |
0.0 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 2 |
Hb_001584_140 |
0.0443903104 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 3 |
Hb_001789_030 |
0.0761053117 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 4 |
Hb_001205_250 |
0.0828991116 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 5 |
Hb_001518_080 |
0.0872789059 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 6 |
Hb_000008_290 |
0.0873582132 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 7 |
Hb_005488_020 |
0.0885691226 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 8 |
Hb_000136_110 |
0.0898190244 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 9 |
Hb_000890_060 |
0.0900877211 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 10 |
Hb_007426_110 |
0.0921289805 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_001655_030 |
0.0925907386 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000915_260 |
0.0937615017 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001981_040 |
0.0943675311 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 14 |
Hb_001205_120 |
0.0944195329 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 15 |
Hb_000441_220 |
0.0952534379 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_015807_050 |
0.0959145755 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 17 |
Hb_079526_040 |
0.0963060966 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 18 |
Hb_002811_270 |
0.096949703 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 19 |
Hb_000163_090 |
0.0974733923 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 20 |
Hb_001188_060 |
0.0975853039 |
- |
- |
- |