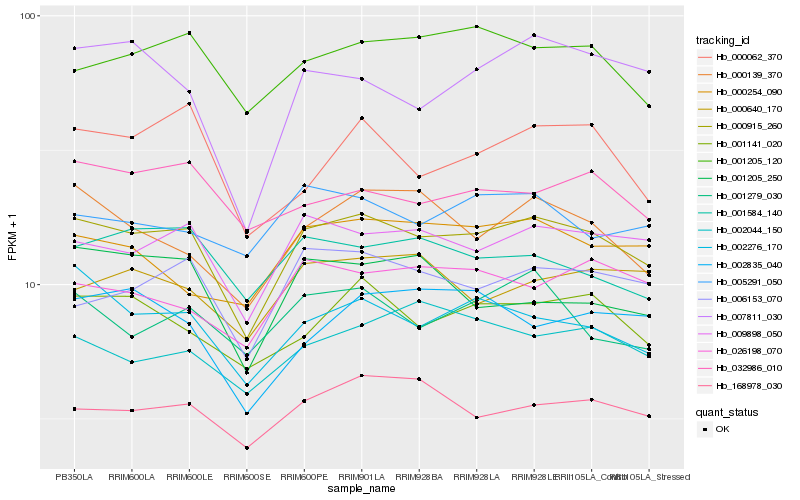
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000915_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009898_050 |
0.0615528475 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 3 |
Hb_002276_170 |
0.0699331485 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001141_020 |
0.0731728574 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 5 |
Hb_168978_030 |
0.0757585913 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 6 |
Hb_006153_070 |
0.0760943097 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001205_120 |
0.0772098562 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 8 |
Hb_000139_370 |
0.0776442865 |
- |
- |
ubiquitin-activating enzyme E1c, putative [Ricinus communis] |
| 9 |
Hb_001584_140 |
0.0818909052 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 10 |
Hb_001279_030 |
0.082199923 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 11 |
Hb_032986_010 |
0.0830395947 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 12 |
Hb_026198_070 |
0.0846552816 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 13 |
Hb_005291_050 |
0.0852663851 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000062_370 |
0.0855518468 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 15 |
Hb_000254_090 |
0.0858709348 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 16 |
Hb_000640_170 |
0.0865744191 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007811_030 |
0.086671901 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_002044_150 |
0.0873938813 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 19 |
Hb_002835_040 |
0.0881322454 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001205_250 |
0.0883809452 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |