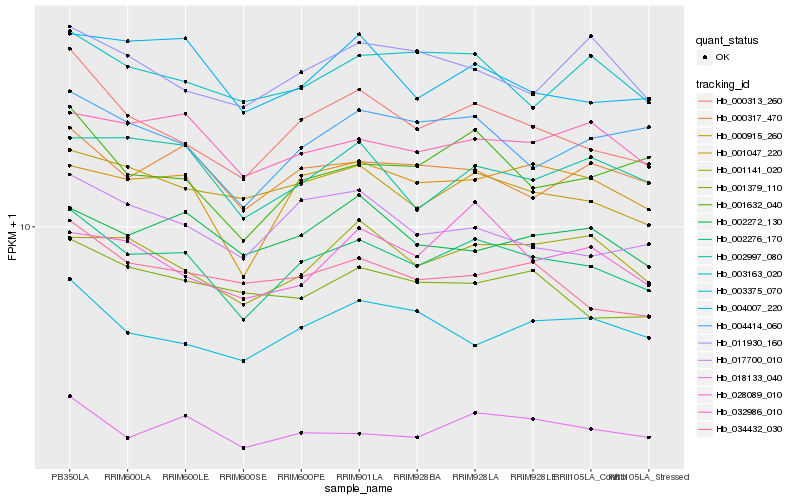
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_170 |
0.0 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000313_260 |
0.0560813329 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 3 |
Hb_000915_260 |
0.0699331485 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001047_220 |
0.0718379934 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 5 |
Hb_000317_470 |
0.0737521625 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 6 |
Hb_001632_040 |
0.0756304915 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_004414_060 |
0.0756993636 |
- |
- |
hypothetical protein JCGZ_17225 [Jatropha curcas] |
| 8 |
Hb_032986_010 |
0.0784212053 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 9 |
Hb_017700_010 |
0.0796897069 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 10 |
Hb_034432_030 |
0.0801268221 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004007_220 |
0.0809933976 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 12 |
Hb_003163_020 |
0.0813415406 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001141_020 |
0.0813709138 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 14 |
Hb_001379_110 |
0.0822854391 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 15 |
Hb_002997_080 |
0.0824203338 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 16 |
Hb_018133_040 |
0.0847053015 |
- |
- |
PREDICTED: probable apyrase 7 [Jatropha curcas] |
| 17 |
Hb_003375_070 |
0.0873421659 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 18 |
Hb_002272_130 |
0.0884912657 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 19 |
Hb_011930_160 |
0.0895778271 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 20 |
Hb_028089_010 |
0.0905465776 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |