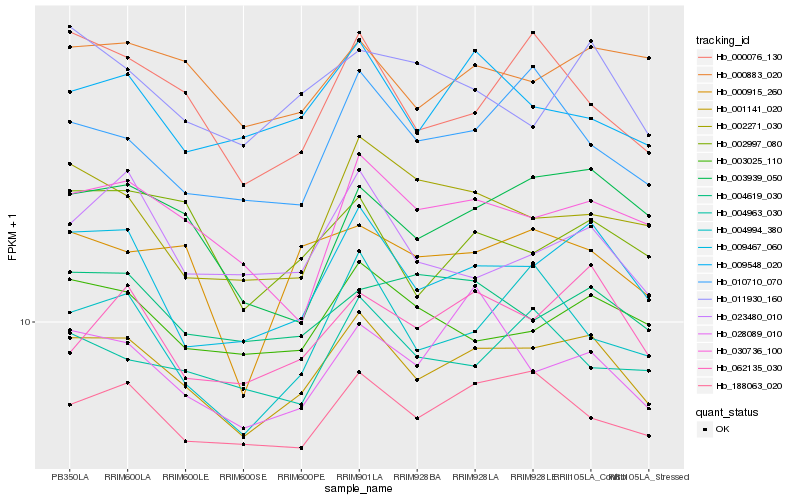
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_020 |
0.0 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 2 |
Hb_009467_060 |
0.051920431 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_188063_020 |
0.0610843294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003025_110 |
0.0661735425 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000883_020 |
0.0695357235 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 6 |
Hb_009548_020 |
0.0705412947 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 7 |
Hb_030736_100 |
0.0717410396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_062135_030 |
0.072170641 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_004619_030 |
0.0727235757 |
- |
- |
PREDICTED: uncharacterized protein LOC105633364 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000915_260 |
0.0731728574 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 11 |
Hb_028089_010 |
0.0748026181 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |
| 12 |
Hb_003939_050 |
0.0752914539 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_011930_160 |
0.0756367283 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 14 |
Hb_004963_030 |
0.0763303477 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000076_130 |
0.0765832113 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 16 |
Hb_002997_080 |
0.0767751483 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 17 |
Hb_010710_070 |
0.0774388333 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_002271_030 |
0.077938649 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 19 |
Hb_023480_010 |
0.0789108601 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_004994_380 |
0.07918324 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |