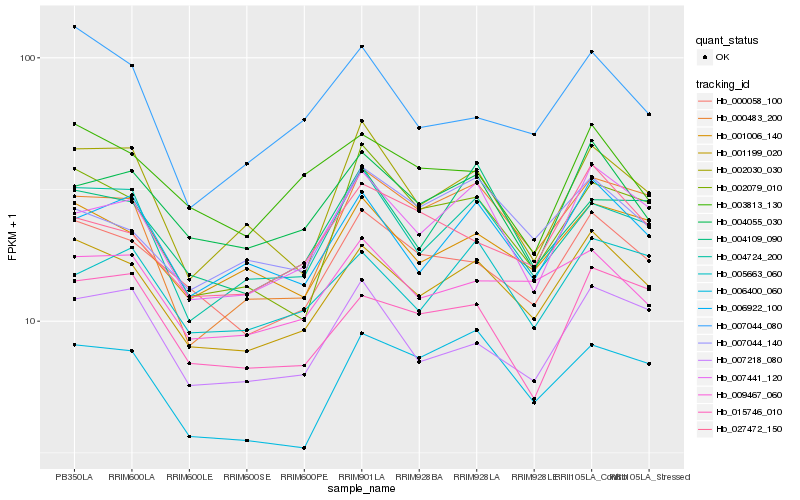
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001199_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 2 |
Hb_000058_100 |
0.0573202515 |
- |
- |
PREDICTED: RNA-directed DNA methylation 4 [Jatropha curcas] |
| 3 |
Hb_001006_140 |
0.062909047 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 19 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004724_200 |
0.0665026581 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 5 |
Hb_007441_120 |
0.0669215366 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 6 |
Hb_006922_100 |
0.0682524761 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004055_030 |
0.0698951103 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 8 |
Hb_009467_060 |
0.0700981793 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006400_060 |
0.0732059751 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007218_080 |
0.0757486704 |
- |
- |
PREDICTED: uncharacterized protein LOC105631523 [Jatropha curcas] |
| 11 |
Hb_004109_090 |
0.0785667917 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_002030_030 |
0.0789865893 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 13 |
Hb_015746_010 |
0.0791412621 |
- |
- |
PREDICTED: triacylglycerol lipase 2-like [Jatropha curcas] |
| 14 |
Hb_005663_060 |
0.0792160572 |
- |
- |
PREDICTED: uncharacterized protein LOC105770600 [Gossypium raimondii] |
| 15 |
Hb_000483_200 |
0.0801601383 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_007044_140 |
0.0806726065 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta [Jatropha curcas] |
| 17 |
Hb_027472_150 |
0.0810290577 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 10 [Jatropha curcas] |
| 18 |
Hb_002079_010 |
0.0811622044 |
- |
- |
hypothetical protein POPTR_0001s01970g [Populus trichocarpa] |
| 19 |
Hb_007044_080 |
0.0823268943 |
- |
- |
PREDICTED: transmembrane protein 33 homolog [Jatropha curcas] |
| 20 |
Hb_003813_130 |
0.082338905 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |