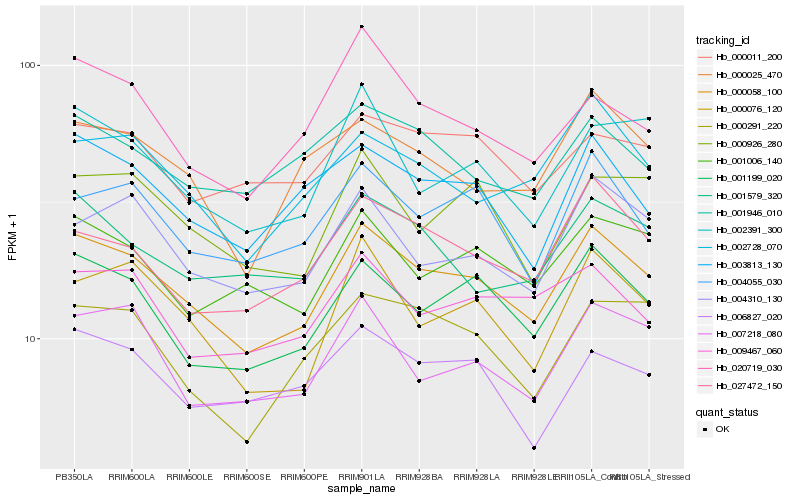
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000058_100 |
0.0 |
- |
- |
PREDICTED: RNA-directed DNA methylation 4 [Jatropha curcas] |
| 2 |
Hb_001199_020 |
0.0573202515 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 3 |
Hb_003813_130 |
0.0694929014 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 4 |
Hb_000076_120 |
0.069599841 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 5 |
Hb_027472_150 |
0.0697350398 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 10 [Jatropha curcas] |
| 6 |
Hb_004055_030 |
0.0700374341 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 7 |
Hb_004310_130 |
0.0707579462 |
- |
- |
PREDICTED: uncharacterized protein LOC105643225 [Jatropha curcas] |
| 8 |
Hb_002728_070 |
0.0722960454 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 9 |
Hb_007218_080 |
0.0731698798 |
- |
- |
PREDICTED: uncharacterized protein LOC105631523 [Jatropha curcas] |
| 10 |
Hb_001006_140 |
0.0743194108 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 19 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000025_470 |
0.075245823 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 12 |
Hb_001579_320 |
0.0756383083 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 13 |
Hb_006827_020 |
0.0762781735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000291_220 |
0.0766105879 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105638855 [Jatropha curcas] |
| 15 |
Hb_000926_280 |
0.0766625287 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 16 |
Hb_001946_010 |
0.0770750286 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_002391_300 |
0.0772962077 |
- |
- |
hypothetical protein PHAVU_002G104100g [Phaseolus vulgaris] |
| 18 |
Hb_000011_200 |
0.0773743658 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009467_060 |
0.0774724409 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_020719_030 |
0.0786719111 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |