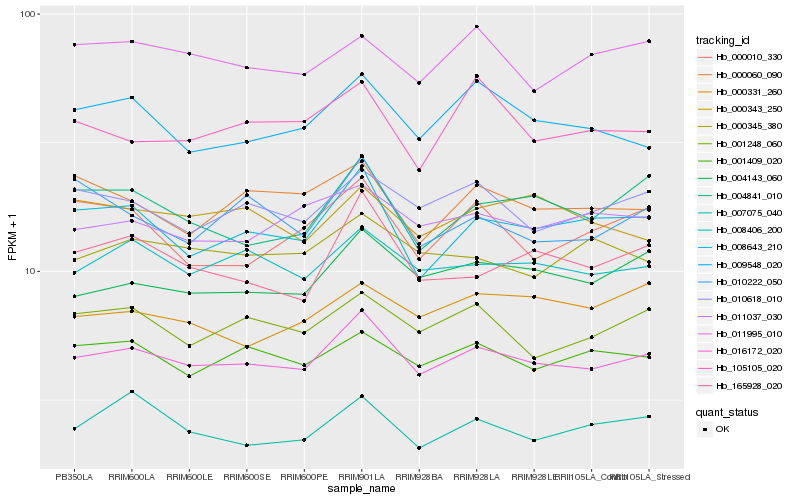
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016172_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_008406_200 |
0.0500050742 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008643_210 |
0.0545004034 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004143_060 |
0.0546590701 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_010618_010 |
0.0561717983 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 6 |
Hb_001409_020 |
0.0583476598 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105638648 [Jatropha curcas] |
| 7 |
Hb_001248_060 |
0.0597566012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011037_030 |
0.0607007676 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 9 |
Hb_009548_020 |
0.0624920786 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 10 |
Hb_165928_020 |
0.0636963993 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000345_380 |
0.0645430456 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 12 |
Hb_004841_010 |
0.0651780245 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000060_090 |
0.0663072892 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a [Jatropha curcas] |
| 14 |
Hb_011995_010 |
0.0686852522 |
- |
- |
dc50, putative [Ricinus communis] |
| 15 |
Hb_000343_250 |
0.0696585735 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 16 |
Hb_007075_040 |
0.0713114045 |
- |
- |
hypothetical protein CISIN_1g039518mg, partial [Citrus sinensis] |
| 17 |
Hb_010222_050 |
0.0721013368 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000331_260 |
0.07253612 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 19 |
Hb_105105_020 |
0.0729272131 |
- |
- |
vacuolar protein sorting vps41, putative [Ricinus communis] |
| 20 |
Hb_000010_330 |
0.073652746 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |