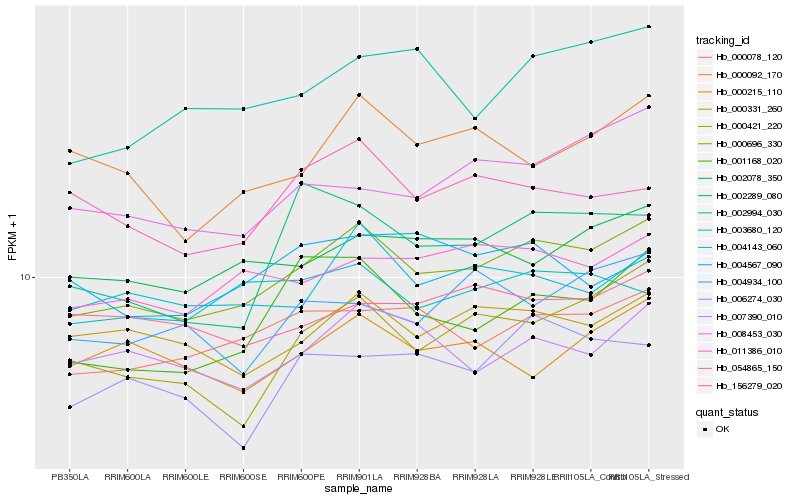
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_330 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647254 [Jatropha curcas] |
| 2 |
Hb_000078_120 |
0.0584180095 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 3 |
Hb_011386_010 |
0.064957231 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 4 |
Hb_004143_060 |
0.0671424994 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002078_350 |
0.0674870704 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 6 |
Hb_000331_260 |
0.0679708858 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001168_020 |
0.068050484 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 8 |
Hb_007390_010 |
0.0698963264 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_002994_030 |
0.0701111845 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 10 |
Hb_000421_220 |
0.0726290125 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_008453_030 |
0.0750664249 |
- |
- |
Thioredoxin superfamily protein [Theobroma cacao] |
| 12 |
Hb_000092_170 |
0.0756948129 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 13 |
Hb_003680_120 |
0.0775091615 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL7 [Jatropha curcas] |
| 14 |
Hb_054865_150 |
0.0781867589 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein Q [Jatropha curcas] |
| 15 |
Hb_156279_020 |
0.0797365584 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 16 |
Hb_006274_030 |
0.0806943627 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 17 |
Hb_004934_100 |
0.0815833005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000215_110 |
0.0817313114 |
- |
- |
PREDICTED: uncharacterized protein C3F10.06c isoform X1 [Jatropha curcas] |
| 19 |
Hb_004567_090 |
0.0819918231 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 20 |
Hb_002289_080 |
0.0820145082 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |