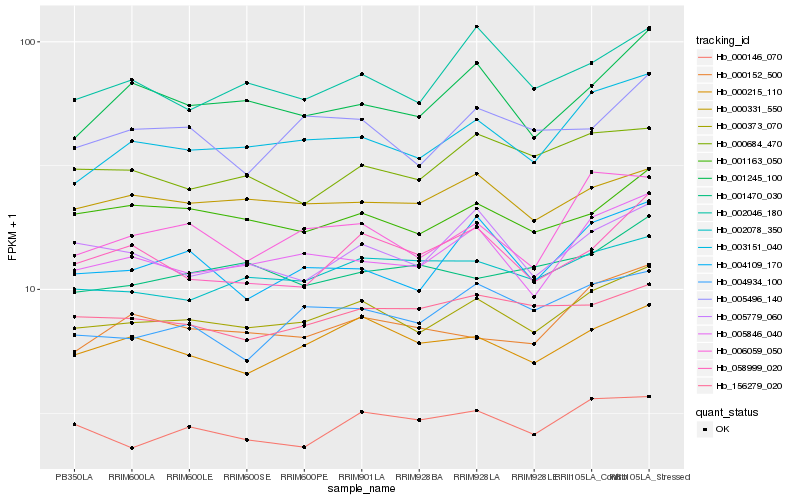
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000373_070 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003151_040 |
0.0507908646 |
transcription factor |
TF Family: Alfin-like |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_000215_110 |
0.0512119438 |
- |
- |
PREDICTED: uncharacterized protein C3F10.06c isoform X1 [Jatropha curcas] |
| 4 |
Hb_005846_040 |
0.051919821 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 4 [Populus euphratica] |
| 5 |
Hb_001163_050 |
0.0524265912 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 6 |
Hb_000331_550 |
0.0540068235 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 7 |
Hb_002078_350 |
0.0572604638 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 8 |
Hb_005779_060 |
0.0606866382 |
- |
- |
PREDICTED: uncharacterized protein LOC105643212 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005496_140 |
0.0619668551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000146_070 |
0.0628512681 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000152_500 |
0.0632961229 |
- |
- |
PREDICTED: probable mitochondrial intermediate peptidase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002046_180 |
0.0635490628 |
- |
- |
PREDICTED: uncharacterized protein LOC105641295 [Jatropha curcas] |
| 13 |
Hb_006059_050 |
0.0635726718 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 14 |
Hb_058999_020 |
0.063670671 |
- |
- |
PREDICTED: mitochondrial outer membrane import complex protein METAXIN [Jatropha curcas] |
| 15 |
Hb_004109_170 |
0.0643546267 |
- |
- |
PREDICTED: probable ribokinase [Jatropha curcas] |
| 16 |
Hb_156279_020 |
0.0643936361 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 17 |
Hb_004934_100 |
0.0647461971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000684_470 |
0.0654475489 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 19 |
Hb_001245_100 |
0.0660850019 |
transcription factor |
TF Family: Alfin-like |
ATP synthase alpha subunit mitochondrial, putative [Ricinus communis] |
| 20 |
Hb_001470_030 |
0.0661306876 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3 isoform X2 [Jatropha curcas] |