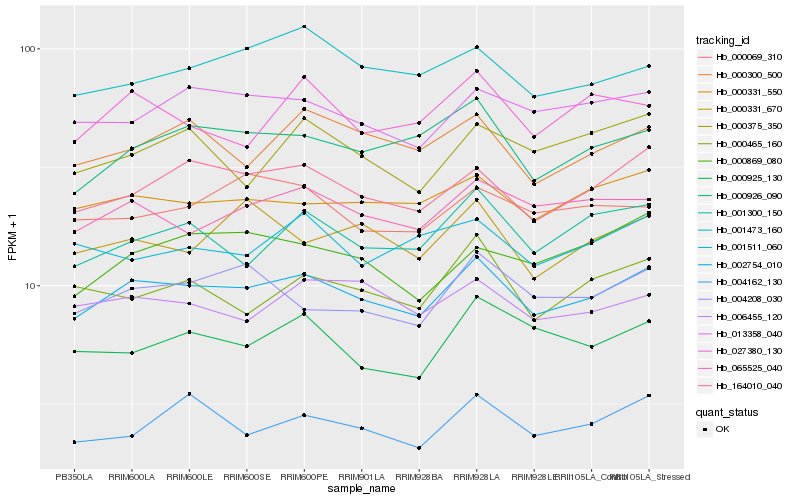
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002754_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 2 |
Hb_164010_040 |
0.0530231897 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000331_550 |
0.0589623097 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 4 |
Hb_000926_090 |
0.0621512538 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_065525_040 |
0.0623841675 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 6 |
Hb_013358_040 |
0.0645714474 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 7 |
Hb_001300_150 |
0.0661446999 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 8 |
Hb_000300_500 |
0.0672514377 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 9 |
Hb_004208_030 |
0.0690906825 |
transcription factor |
TF Family: MYB-related |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 10 |
Hb_006455_120 |
0.0702836838 |
- |
- |
n6-DNA-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001473_160 |
0.0720496402 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 12 |
Hb_000465_160 |
0.0731081694 |
- |
- |
PREDICTED: uncharacterized protein LOC105632297 [Jatropha curcas] |
| 13 |
Hb_000869_080 |
0.0742174041 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 14 |
Hb_027380_130 |
0.0756380756 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000069_310 |
0.0765961865 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |
| 16 |
Hb_000925_130 |
0.0772569855 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000375_350 |
0.0772802145 |
- |
- |
PREDICTED: uncharacterized protein LOC105641632 [Jatropha curcas] |
| 18 |
Hb_001511_060 |
0.0776876106 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 19 |
Hb_004162_130 |
0.0776950034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000331_670 |
0.0777079449 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |