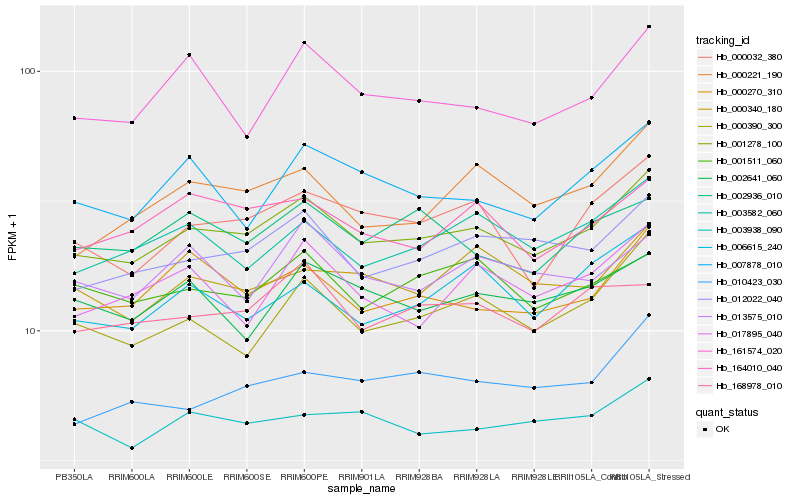
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001278_100 |
0.0 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 2 |
Hb_012022_040 |
0.0523573805 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 3 |
Hb_000032_380 |
0.0600933914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000270_310 |
0.0606473305 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 5 |
Hb_003582_060 |
0.0613971883 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000390_300 |
0.063171188 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 7 |
Hb_164010_040 |
0.0639816552 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000340_180 |
0.0645847521 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of NFKB 1 [Jatropha curcas] |
| 9 |
Hb_003938_090 |
0.0648505147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000221_190 |
0.0648932037 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 11 |
Hb_001511_060 |
0.0658934725 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 12 |
Hb_007878_010 |
0.0661491013 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 13 |
Hb_168978_010 |
0.0662568025 |
- |
- |
PREDICTED: uncharacterized protein LOC105630751 [Jatropha curcas] |
| 14 |
Hb_002641_060 |
0.0669983553 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 15 |
Hb_010423_030 |
0.0678954229 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 16 |
Hb_161574_020 |
0.0681721941 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_006615_240 |
0.0690490776 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 18 |
Hb_002936_010 |
0.0707515632 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 19 |
Hb_017895_040 |
0.0710096094 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 20 |
Hb_013575_010 |
0.0710752468 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |