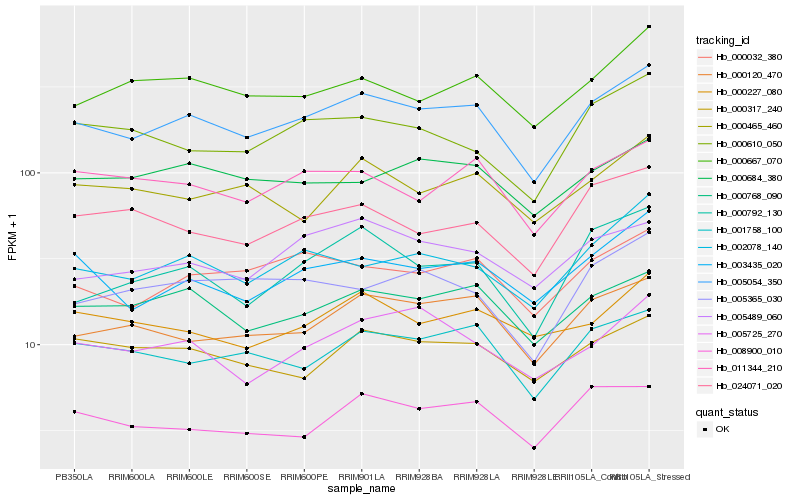
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_350 |
0.0 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P0 [Vitis vinifera] |
| 2 |
Hb_000120_470 |
0.0689910629 |
- |
- |
hypothetical protein VITISV_017372 [Vitis vinifera] |
| 3 |
Hb_000768_090 |
0.0700155045 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Jatropha curcas] |
| 4 |
Hb_003435_020 |
0.0720802385 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001758_100 |
0.0729908708 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000032_380 |
0.073627471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002078_140 |
0.0784546702 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 8 |
Hb_011344_210 |
0.0812126567 |
- |
- |
PREDICTED: probable calcium-binding protein CML13 [Jatropha curcas] |
| 9 |
Hb_000667_070 |
0.0817962825 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 10 |
Hb_005489_060 |
0.0820289872 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 11 |
Hb_000792_130 |
0.0841447407 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 12 |
Hb_000317_240 |
0.0846443226 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 13 |
Hb_008900_010 |
0.0851717933 |
- |
- |
PREDICTED: uncharacterized protein LOC105773666 [Gossypium raimondii] |
| 14 |
Hb_024071_020 |
0.0859487529 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 15 |
Hb_000610_050 |
0.0860262979 |
- |
- |
skp1, putative [Ricinus communis] |
| 16 |
Hb_005725_270 |
0.0867572828 |
- |
- |
PREDICTED: uncharacterized protein LOC105630802 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000684_380 |
0.0869389117 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 18 |
Hb_005365_030 |
0.0873363302 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 19 |
Hb_000465_460 |
0.0874143223 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 20 |
Hb_000227_080 |
0.0874740693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |