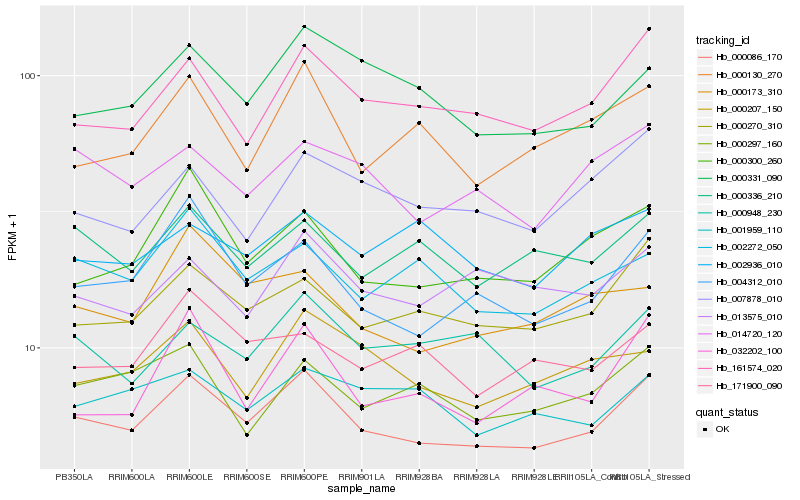
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_170 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 2 |
Hb_000270_310 |
0.0609586342 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 3 |
Hb_004312_010 |
0.0685024248 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 4 |
Hb_161574_020 |
0.0685235528 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 5 |
Hb_014720_120 |
0.0729013886 |
- |
- |
Phosphoenolpyruvate/phosphate translocator 1, chloroplastic -like protein [Gossypium arboreum] |
| 6 |
Hb_000336_210 |
0.0739637874 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002272_050 |
0.0753009203 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 8 |
Hb_000300_260 |
0.0754888208 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 9 |
Hb_000948_230 |
0.0766371644 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_000207_150 |
0.0772910551 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 11 |
Hb_000297_160 |
0.0785249761 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 12 |
Hb_000331_090 |
0.0786825 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 13 |
Hb_001959_110 |
0.0794504661 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 14 |
Hb_013575_010 |
0.0796659023 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 15 |
Hb_171900_090 |
0.0805487927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007878_010 |
0.0819436853 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 17 |
Hb_000130_270 |
0.0822742533 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000173_310 |
0.0827258366 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 19 |
Hb_032202_100 |
0.0847920568 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 20 |
Hb_002936_010 |
0.0859772409 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |