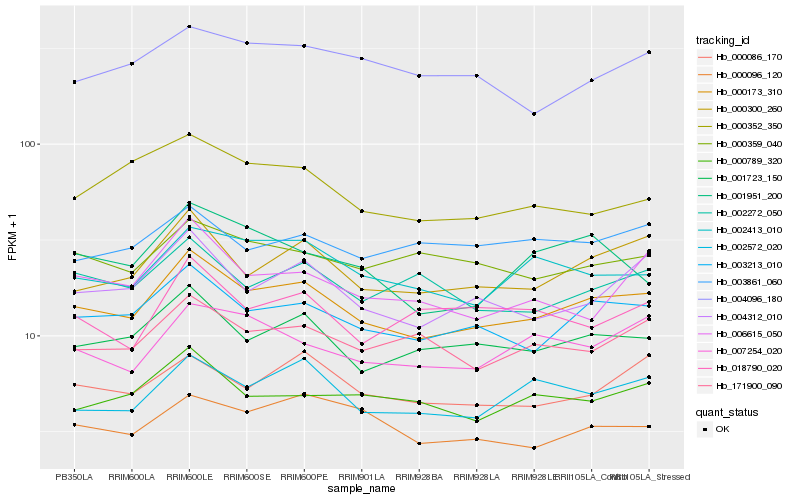
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003213_010 |
0.0533362463 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 3 |
Hb_001723_150 |
0.0639969416 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000300_260 |
0.0640018551 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 5 |
Hb_004312_010 |
0.0729457788 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002572_020 |
0.0753970643 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 7 |
Hb_002413_010 |
0.0759425005 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_171900_090 |
0.0788876392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007254_020 |
0.0820067278 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 10 |
Hb_000789_320 |
0.0825481173 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000086_170 |
0.0827258366 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 12 |
Hb_002272_050 |
0.0850484451 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 13 |
Hb_000096_120 |
0.0862285081 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 14 |
Hb_000359_040 |
0.0864578926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006615_050 |
0.0884513615 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 16 |
Hb_000352_350 |
0.0917710495 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004096_180 |
0.0920821799 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 18 |
Hb_001951_200 |
0.0926082956 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_018790_020 |
0.0927367284 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 20 |
Hb_003861_060 |
0.0929371663 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |