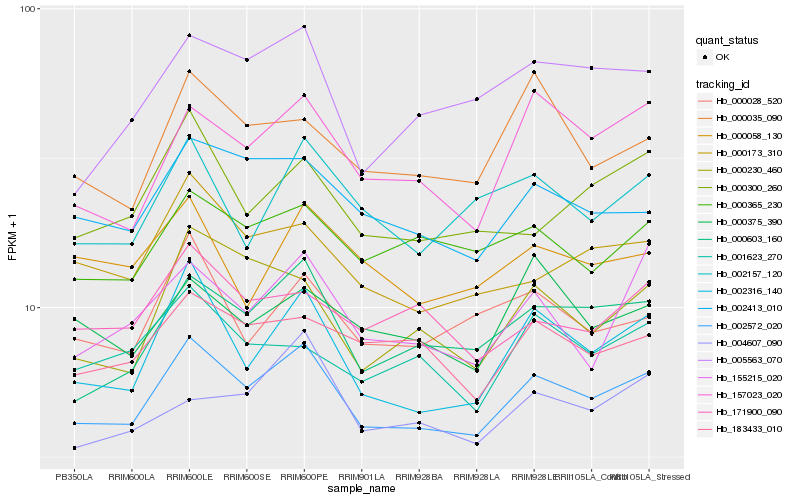
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002572_020 |
0.0 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 2 |
Hb_000603_160 |
0.0611701141 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 3 |
Hb_002316_140 |
0.0657189283 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 4 |
Hb_002413_010 |
0.0677503808 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000365_230 |
0.0733852253 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 6 |
Hb_000173_310 |
0.0753970643 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 7 |
Hb_183433_010 |
0.0755616738 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 8 |
Hb_157023_020 |
0.0757449436 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000028_520 |
0.0787495551 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 10 |
Hb_002157_120 |
0.0804542363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_171900_090 |
0.0813508798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_155215_020 |
0.0831486445 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 13 |
Hb_000300_260 |
0.0832844282 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 14 |
Hb_000035_090 |
0.0832911761 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 15 |
Hb_004607_090 |
0.0833958628 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 16 |
Hb_001623_270 |
0.083625737 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 17 |
Hb_000230_460 |
0.0842989351 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 18 |
Hb_005563_070 |
0.0844189283 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 19 |
Hb_000375_390 |
0.0844253516 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 20 |
Hb_000058_130 |
0.0847775872 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |