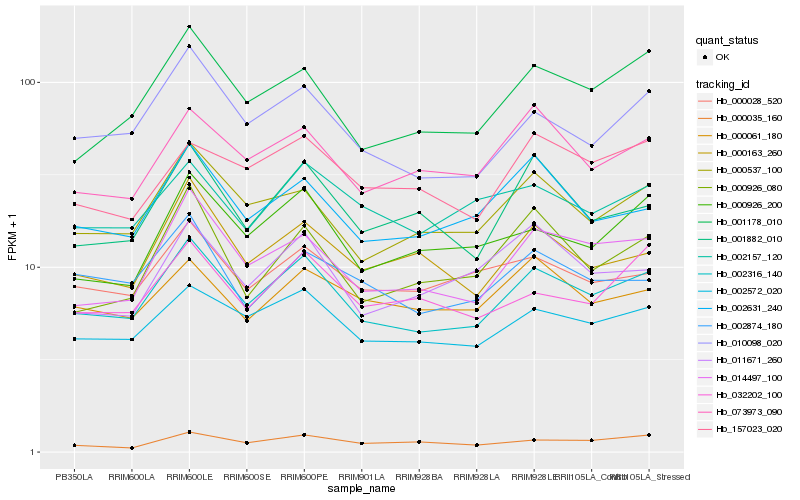
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002316_140 |
0.0 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 2 |
Hb_002572_020 |
0.0657189283 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 3 |
Hb_010098_020 |
0.0706003225 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_014497_100 |
0.0709439334 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000537_100 |
0.0724742811 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001178_010 |
0.0795650884 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 7 |
Hb_073973_090 |
0.085641914 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 8 |
Hb_002631_240 |
0.0862287094 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 9 |
Hb_000926_200 |
0.0885732113 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000028_520 |
0.0896337385 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 11 |
Hb_002157_120 |
0.0908667399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002874_180 |
0.0917737906 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001882_010 |
0.0918198504 |
- |
- |
- |
| 14 |
Hb_000163_260 |
0.0925934867 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 15 |
Hb_032202_100 |
0.0943427551 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 16 |
Hb_157023_020 |
0.0947001679 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000061_180 |
0.0957803776 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 18 |
Hb_000035_160 |
0.0967928119 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 19 |
Hb_011671_260 |
0.0980259422 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000926_080 |
0.0991842177 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |